|

CLICK ON weeks 0 - 40 and follow along every 2 weeks of fetal development
|
||||||||||||||||||||||||||||
Editing mice to study human leukemia Using multiplex CRISPR-Cas9 editing of human hematopoietic, blood-forming stem cells transplanted into mice, a team of scientists from the Broad Institute of MIT, Harvard Medical School, and the Dana-Farber Cancer Institute (DFCI) designed customized mouse models in order to study the progression of leukemia. In a number of experiments, their animal models successfully reflected human responses to a therapeutic agent commonly used to treat blood cancers. "With our models, we can really test - in a very controlled fashion, in the right setting, and using the right cells - the genetic predictors of response to specific agents." The research team started by examining large-scale sequencing data from Dana Ebert's lab at the Department of Medical Oncology, Dana Farber Cancer Institute, and from The Cancer Genome Atlas, to determine which combinations of mutations occur most commonly in myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML), blood cancers in which the bone marrow fails to produce healthy blood cells. The researchers landed on nine genes that are recurrently mutated in MDS and AML. "We use human genetics to teach us which combinations of mutations lead to cancer," explained Ebert. "If we have sequencing data from enough tumors, we can identify the genes that are mutated recurrently and which combinations of mutations co-occur more commonly than expected by chance." Currently, many cancer model systems (such as cell lines) do not reflect cancer genetics a particular investigator wants to study, often leaving both researchers and patients at a disadvantage. One strategy is to transplant an actual human cancer sample into a mouse, but tissue often doesn't engraft well. Researchers are also only able to test against specific combinations of mutations in a given cancer sample in the first place. So the team developed a pipeline to insert specific MDS-driving mutation combinations into new lab models. The work appears in Cell Stem Cell Tothova:"Say we're trying to develop a new drug against a particular combination of mutations, which we know about through our cancer sequencing efforts. You might not have any sample available to study with that particular combination of mutations. We wanted to be able to engineer the right lesions in human cells, let them expand in mice, and generate an accurate genetic model of disease for testing new therapies. This has been a longstanding goal for cancer researchers, and for the pharmaceutical industry, for a very long time." To create models with the right mutations, Tothova and her team established a customizable system to introduce cancer-driving mutations into human hematopoietic stem cells, where MDS and AML originate. They already had extensive experience working with hematopoietic stem cells and progenitor cells, taken from umbilical cord blood or adult bone marrow. In 2014, they published a Nature Biotechnology paper describing using the CRISPR-Cas9 system to create similar models of mouse cancers. This time, the team aimed to model MDS in human cells, a much more challenging goal. Taking primary cells from healthy donors and using CRISPR-Cas9, they engineered a number of different mutation combinations, rather than one single alteration. These reflected the actual complexity of tumor mutations seen in patients. "Nobody so far has done this kind of multiplex CRISPR engineering in the actual hematopoietic stem cell compartment, adding specific mutations in combination to generate disease models." From there, the team injected the stem cells into the mice circulatory systems, where a portion incorporated into the bone marrow. Monitoring their progression, scientists extracted and sequenced the human cells five months later to determine which engineered cells were successfully propagated, and which mutations became the most common over time in pre-malignant and early malignant states. "We are able to recapitulate findings previously seen in human clinical trials, which makes us feel more confident in the power of these models," said Tothova. "The data that comes from patients reflects the most important experiment we are trying to understand." She is currently working with clinical collaborators at DFCI to extend some of these findings into clinical trials. The team believes their approach to create this type of leukemia progression model for therapeutic testing can be applied to other types of cancer as well, as long as sequencing data is available to choose appropriate mutations and progenitor cells can be acquired from the desired tissue. "People in the field are hungry for these kinds of models," said Ebert. "We are modeling the disease in the right cellular context with a genetic complexity that reflects what we see in patients. This hasn't been done before, and it could become a really beneficial tool." Highlights • CD34+ cells can be edited with multiple tumor suppressor lesions with >70% efficiency • Edited CD34+ cells expand in vivo after transplantation into immunodeficient mice • CD34+ cells engineered with combinations of mutations model clonal dynamics in vivo • Engineered cells model genotype-specific therapeutic liabilities of myeloid neoplasia Summary Hematologic malignancies are driven by combinations of genetic lesions that have been difficult to model in human cells. We used CRISPR/Cas9 genome engineering of primary adult and umbilical cord blood CD34+ human hematopoietic stem and progenitor cells (HSPCs), the cells of origin for myeloid pre-malignant and malignant diseases, followed by transplantation into immunodeficient mice to generate genetic models of clonal hematopoiesis and neoplasia. Human hematopoietic cells bearing mutations in combinations of genes, including cohesin complex genes, observed in myeloid malignancies generated immunophenotypically defined neoplastic clones capable of long-term, multi-lineage reconstitution and serial transplantation. Employing these models to investigate therapeutic efficacy, we found that TET2 and cohesin-mutated hematopoietic cells were sensitive to azacitidine treatment. These findings demonstrate the potential for generating genetically defined models of human myeloid diseases, and they are suitable for examining the biological consequences of somatic mutations and the testing of therapeutic agents. Authors: Zuzana Tothova, John M. Krill-Burger, Katerina D. Popova, Catherine C. Landers, Quinlan L. Sievers, David Yudovich, Roger Belizaire, Jon C. Aster, Elizabeth A. Morgan, Aviad Tsherniak, Benjamin L. Ebert This project was supported in part by funding from the Broad Institute's BroadIgnite program, which connects rising philanthropists to emerging scientists pursuing high-risk -- and potentially high-reward -- ideas. Additional funding was provided by the NIH (R01HL082945, P01CA108631, 5K12CA087723-12, T32GM007753, and DK106829), the Edward P. Evans Foundation, the Gabrielle's Angel Foundation, the LLS Scholar Award, the LLS Special Fellow Award, the ASCO Young Investigator Award, and the ASH/EHA Translational Research in Hematology Award. Return to top of page |
Oct 12, 2017 Fetal Timeline Maternal Timeline News News Archive 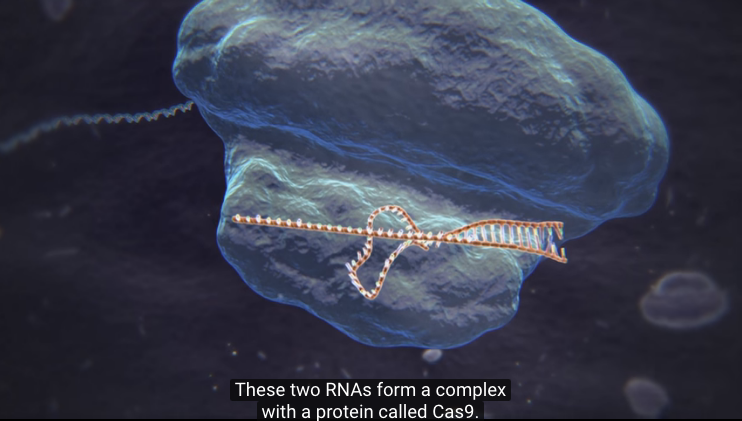 CRISPR-Cas achieves its target via a small RNA that can easily be swapped for another RNA target in a new site. There are a number of different types of CRISPR-Cas systems in nature. The CRISPR-Cas9 system uses just a single protein, Cas9, to find and destroy target DNA. Image Credit: Broad Institute
|
||||||||||||||||||||||||||||

