|

CLICK ON weeks 0 - 40 and follow along every 2 weeks of fetal development
|
||||||||||||||||||||||||||||
A gene initiating neuron formation may lead to repair University of California Los Angeles (UCLA) researchers have discovered the inner workings of a gene network that regulates the development of spinal motor neurons in the growing chicken and mouse embryo. During embryonic development of the spinal cord, different types of nerve cells form from precursor cells, called neural progenitor cells. This research answers a long-standing question about why motor neurons (MNs) — the spinal cord nerve cells that control muscle movement — form much faster than other neurons. Analysis suggests that a gene called Olig2 produces a protein only expressed in neural progenitor cells which create motor neurons. Olig2 promotes motor neuron formation by interfering with the activity of Hes genes that supress neuron development. The study was published in the journal PLOS Biology by co-senior author Bennett Novitch, a member of the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research at UCLA, along with his collaborators from the Francis Crick Institute in London, UK. The research has implications for facilitating the laboratory production of motor neurons from stem cells. Stem cell-derived motor neurons could then be used to repair diseased or damaged spinal cords and expand the study of neurodegenerative diseases such as amyotrophic lateral sclerosis (also known as Lou Gehrig's disease) and spinal muscular atrophy. "This study provides an unprecedented detailed view of how embryos produce the different cell types found in a mature spinal cord. The new experimental techniques we used, combined with collaborative efforts of biologists and computer scientists, are allowing us to gain new insight into the exquisite accuracy of embryo development." It's been known for a long time that different types of neurons form at differing speeds in a developing embryo, with MNs forming faster than other types of nerve cells that populate the spinal cord. However, researchers didn't know why or how MNs develop. So the research team used the latest molecular technique that assesses gene activity. Using an approach called single cell transcriptional profiling, which allows the activity of all genes in individual cell to be measured simultaneously, the team took snapshots of gene activity in about 200 cells in the process of becoming motor neurons. They then analyzed this data using custom computer software they developed, and reconstructed how gene activity changes MN formation. Their work confirmed increased Olig2 essentially establishes MN progenitor cell differentiation in the prescence of Sonic Hedgehog (Shh) signalling. It also suppresses Notch signals and the expression of Hes genes that negatively regulate neuronal differentiation. "During embryonic development, the nervous system is constructed following a precise blueprint, with key parts forming at precise times and places, and appropriate numbers. Our research sheds light into how this process is orchestrated specifically for motor neuron development," explains Novitch. Abstract During tissue development, multipotent progenitors differentiate into specific cell types in characteristic spatial and temporal patterns. We addressed the mechanism linking progenitor identity and differentiation rate in the neural tube, where motor neuron (MN) progenitors differentiate more rapidly than other progenitors. Using single cell transcriptomics, we defined the transcriptional changes associated with the transition of neural progenitors into MNs. Reconstruction of gene expression dynamics from these data indicate a pivotal role for the MN determinant Olig2 just prior to MN differentiation. Olig2 represses expression of the Notch signaling pathway effectors Hes1 and Hes5. Olig2 repression of Hes5 appears to be direct, via a conserved regulatory element within the Hes5 locus that restricts expression from MN progenitors. These findings reveal a tight coupling between the regulatory networks that control patterning and neuronal differentiation and demonstrate how Olig2 acts as the developmental pacemaker coordinating the spatial and temporal pattern of MN generation. Author summary How are the right types of cells produced in the right place, at the right time and in the correct numbers, in a developing tissue? One example of where progress has been made towards answering this question is the embryonic spinal cord. In this tissue, extracellular signals, such as the morphogen sonic hedgehog (Shh), control the pattern of generation of molecularly distinct sets of neural progenitors, from which different classes of motor neurons and interneurons are generated. Motor neurons differentiate at a much higher rate than the adjacent interneurons, and this ensures that more motor neurons than interneurons are generated and in an appropriate temporal sequence. To understand the mechanisms responsible for this phenomenon, we investigated the dynamics of the Shh-controlled gene regulatory network, operating as motor neurons form from progenitors. We used these data to uncover the accompanying regulatory mechanisms, and this identified two functions for the transcription factor Olig2. First, Olig2 is essential for establishing motor neuron progenitor identity downstream of Shh signaling. Subsequently, Olig2 directly promotes neuronal differentiation in motor neuron progenitors by suppressing the expression of Hes genes, negative regulators of neuronal differentiation. Together, our findings reveal a tight coupling between the genetic networks that control patterning and neuronal differentiation in motor neuron progenitors and thereby explain their characteristic early and rapid rate of neuronal differentiation. All authors: Andreas Sagner, Zachary B. Gaber, Julien Delile, Jennifer H. Kong, David L. Rousso, Caroline A. Pearson, Steven E. Weicksel, Manuela Melchionda, S. Neda Mousavy Gharavy, James Briscoe, Bennett G. Novitch Search terms: Olig2, tissue development, gene expression, Notch signaling pathway, single cell transcriptomics The authors have no conflicts of interest or financial disclosures relevant to this manuscript. The research was funded by the Wellcome Trust (grant number FC001051; WT098326MA). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. EMBO Long-term fellowship (grant number 1438-2013). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Cancer Research UK (grant number FC001051). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Whitehall Foundation (grant number 2004-05-90-APL). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. March of Dimes Foundation (grant number 5-FY06-7). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. NINDS (grant number R01NS053976, R01NS072804, R01NS085227). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. UK Medical Research Council (grant number FC001051). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Human Frontiers Science Program (grant number LT000401/2014-L). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. People Programme (Marie Curie Actions) of the European Union's Seventh Framework Programme (grant number FP7-2013 624973). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Return to top of page | Feb 6, 2018 Fetal Timeline Maternal Timeline News News Archive 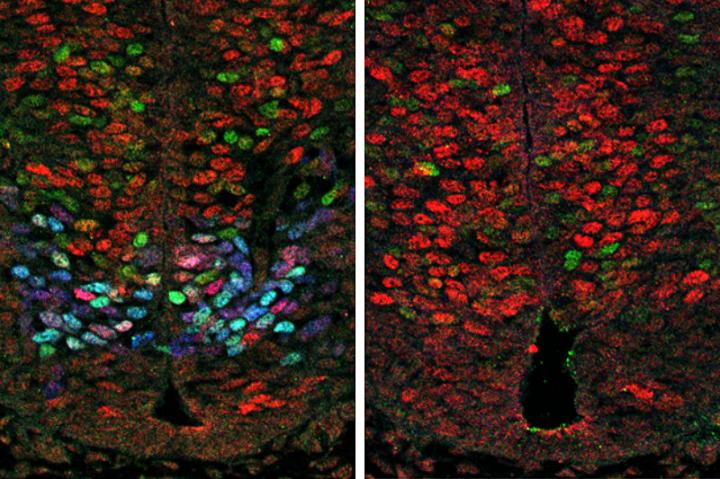 Left: Olig2 expression (blue) results in reduced Hes gene expression (red) and increased motor neuron formation (green). Right: in the absence of Olig2, Hes expression increases and motor neuron formation decreases.
CREDIT
PLOS Biology/UCLA Broad Stem Cell Research Center
|
||||||||||||||||||||||||||||