|
|
Developmental Biology - CRISPR
A New Way to Use CRISPR
New method uses CRISPR technology for tighter gene regulation...
A team of engineers at the University of Delaware (UD) has developed a new way to make use of CRISPR/Cas9 technology. It works by setting off a cascade of cellular activities in a phenomenon known as conditional gene regulation. This new CRISPR function is described in the journal Nature Chemical Biology.
Gene editing with CRISPR technology has been called "one of the biggest science stories of the decade" for its applications to medicine, agriculture and more. CRISPR allows scientists to precisely target and edit DNA within living cells, and could help correct anomalies that cause inherited diseases. In China, the first clinical human trials are even underway.
However, scientists haven't figured out how to program CRISPR systems to target DNA and at the same time integrate information from within the cells being manipulated. Now, UD Professor of Chemical Engineering, Wilfred Chen along with graduate student Ka-Hei Siu, have designed toehold-gated gRNA or thgRNA a hairpin shaped structure that can target and regulate genes in E. coli bacteria.
In traditional CRISPR/Cas9 genome editing, scientists use a single-strand of ribonucleic acid (RNA) to guide the Cas9 enzyme towards the targeted DNA. But Chen and Siu instead install a hairpin-like structure to block RNA from recognizing the target DNA. The hairpin-like structure, or toehold, is exposed and capable of attaching to another RNA. In this way, Chen and Siu can use RNA from within the cell as a trigger switch to open up the hairpin structure and use it as a blocking mechanism, the Cas9 protein binds to it and then seeks the DNA target.
"We want to be able to use this native cellular response to modulate CRISPR/Cas9 protein function and developed a controlled mechanism. Toehold technology offers a plug and play design."
Wilfred Chen PhD, Department of Chemical and Biomolecular Engineering, University of Delaware, Newark, DE, USA
Abstract
Predictable control over gene expression is essential to elicit desired synthetic cellular phenotypes. Although CRISPR–Cas9 offers a simple RNA-guided method for targeted transcriptional control, it lacks the ability to integrate endogenous cellular information for efficient signal processing. Here, we present a new class of riboregulators termed toehold-gated gRNA (thgRNA) by integrating toehold riboswitches into sgRNA scaffolds, and demonstrate their programmability for multiplexed regulation in Escherichia coli with minimal cross-talks.
Authors
Ka-Hei Siu and Wilfred Chen.
Anthony Swerdlow DM, DSc, PhD, and Minouk Schoemaker PhD, scientists at the Institute of Cancer Research, London, co-led the study with Sandler and Nichols.
Acknowledgements
This work was supported by grants to W.C. from the National Science Foundation (MCB1615731 and MCB1817675). We thank D. Liu (Harvard University), T. Pederson (University of Massachusetts Medical School), and M. Koffas (Rensselaer Polytechnic Institute) for their generous gifts of plasmids as noted in the manuscript.
Return to top of page
| |
|
Dec 19, 2018 Fetal Timeline Maternal Timeline News News Archive
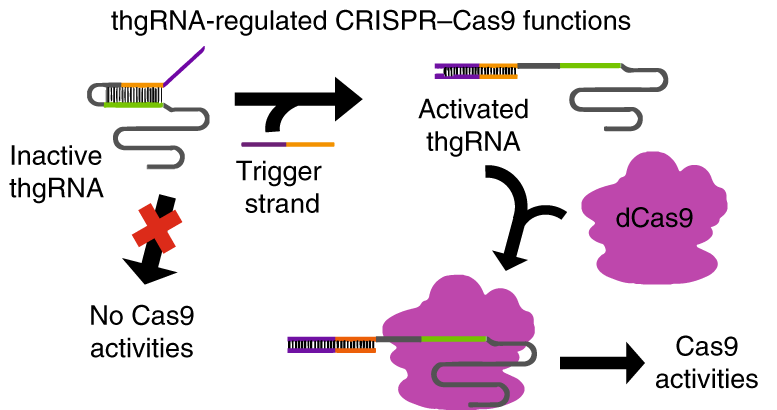 Traditionally CRISPR/Cas9 targets a section of single-stranded RNA guiding the Cas9 enzyme to a chosen DNA strand. Toehold binds to selected parts of the RNA, activating Cas9 to bind and regulate a more closely chosen section of DNA.
|



