|

CLICK ON weeks 0 - 40 and follow along every 2 weeks of fetal development
|
||||||||||||||||||||||||||||
|
Developmental Biology - DNA and Aging True age detected in DNA from blood Specifically, Neretti's group found nucleosomes - a segment of DNA wrapped around a protein core - were spaced consistently in DNA of volunteers in their 20s, but less regularly in older volunteers, especially in unhealthy centenarians. Incredibly, the signal from nucleosome spacing in healthy centenarians was more similar to the signal from people in their 20s than people in their 70s. Nucleosome packing is one aspect of the epigenome in a collection of heritable changes that affect gene expression or activity — without affecting the DNA sequence of the genome. "Among other traits, healthy centenarians preserve the epigenomic profile of younger individuals. As with anything in aging, many things work together, and it is not clear what the cause or the effect of each is. With our cfDNA test, we hope to gain understanding of these epigenetic changes and what they mean." Message in a bottle Scientists first found cfDNA in blood from cancer patients, and that these fragments can be useful for diagnosing cancer. Earlier research had found that cfDNA is produced by dying cells — as the cells die, DNA is cut in between nucleosome packages, according to Neretti. The team at Brown used next-generation sequencing of cfDNA combined with complex computational analysis to reconstruct the pattern made by nucleosome spacing in different regions of the genome - both in areas that are typically open for expressing genes and in areas which are normally tightly packed and therefore unable to be expressed. cfDNA extraction and sequencing processes were developed in collaboration with Ana Maria Caetano Faria from the Universidade Federal de Minas Gerais in Brazil. "cfDNA is somewhat like a message in a bottle that captures what the cell looked like, epigenetically speaking, before it died. A lot of cellular machinery is involved in maintaining nucleosome spacing, and these components can go downhill as you age. However, he added, changes in nucleosome packing produce changes in the accessibility of different parts of the genome, which leads to even more things going awry, including the freeing of normally locked-down genetic elements called transposons. The team did detect a reduction in cfDNA signals at the beginning of two common transposons with increasing age. This suggests that these transposons are less locked-down in the unhealthy centenarians and people in their 70s and thus more likely to be "copying and pasting" themselves into the genome, causing genetic mayhem. Future work The study only analyzed the cfDNA of 12 individuals from Bologna, Italy - three from each group. The samples were collected by collaborator Claudio Franceschi, from the University of Bologna. A larger study is needed to gain the information necessary to use these epigenetic markers to predict biological age, Neretti adds. However, because the cfDNA test uses easy to collect blood instead of invasive tissue samples, he thinks it will be a straight forward process to expand this initial proof-of-concept study. "Ideally, you would like to track a population of individuals over 20 or 30 years to see how each individual's epigenome changes, and the rate of change, as they age," says Neretti. He feels a larger study could allow the association of epigenomic differences with health conditions, lifestyles and/or diets. Meanwhile... The research team continues to refine the test, working to optimize the process of extracting cfDNA from blood. In mice, they can reliably get the amount of cfDNA they need from a quarter teaspoon of blood. Neretti believes they won't need to sequence an entire genome to detect age and health related epigenetic changes. For this study, they did whole-genome sequencing, but in the future he expects sequencing 2 to 5 percent of the genome will be sufficient. In addition to refining their analysis of the positions of nucleosomes, they would like to study another kind of epigenetic marker - DNA methylation patterns in cfDNAs. This might give additional information indicating which tissues the cfDNA was taken from; determine the sources of cfDNA in different ages groups; and which tissues experience more cell death. All of which would provide insights into the aging process. Better understanding the epigenetic changes of aging would aid in developing treatments for age-associated disorders — or someday might be used to determine whether your body is aging faster or slower than typical. Abstract Cell-free (cfDNA) is present in the circulating plasma and other body fluids and is known to originate mainly from apoptotic cells. Here, we provide the first in vivo evidence of global and local chromatin changes in human aging by analyzing cfDNA from the blood of individuals of different age groups. Our results show that nucleosome signals inferred from cfDNA are consistent with the redistribution of heterochromatin observed in cellular senescence and aging observed in other model systems. In addition, we detected a relative cfDNA loss at several genomic locations, such as transcription start and termination sites, 5'UTR of L1HS retrotransposons and dimeric AluY elements with age. Our results also revealed age and deteriorating health status correlate with increased enrichment of signals from cells in different tissues. In conclusion, our results show that the sequencing of circulating cfDNA from human blood plasma can be used as a noninvasive methodology to study age-associated changes to the epigenome in vivo. Authors First author of the paper is Yee Voan Teo, doctoral student in molecular biology, cell biology and biochemistry at Brown University. In addition to Ana Maria Caetano Faria and Claudio Franceschi, other authors on the paper include Miriam Capri, Cristina Morsiani and Grazia Pizza from the University of Bologna. Acknowledgements The National Institutes of Health (grant R01-AG050582) and the Brown-Brazil Collaborative Research Fund (grant GFT640009) supported the research. Return to top of page | Jan 9, 2019 Fetal Timeline Maternal Timeline News News Archive 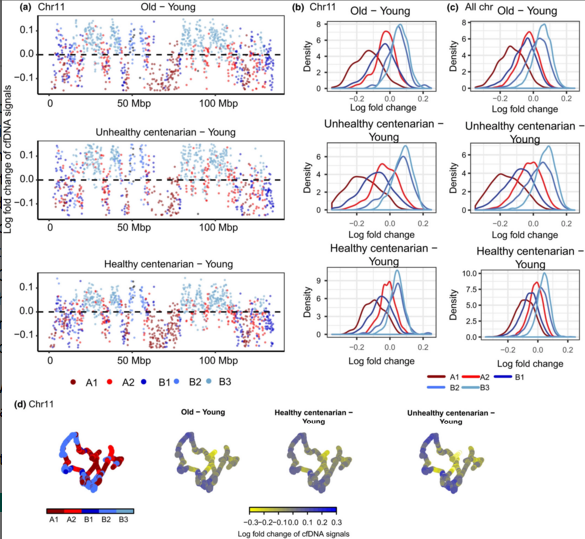 cell-free DNA (cfDNA) signals reveal a loss in compartment A1 - B1 and a gain of signals in compartment B2 - B3 with age. (A) cfDNA signal Log fold changes across chr 11 of each age group relative to young. Colors represent compartments with 100-kb spanning regions based on GM12878 Hi-C data. (B) Distribution of cfDNA signals log fold change by compartments in chr 11. (C) Distribution of log fold change of cfDNA signals by compartments in all chromosomes. (D) 3D organization of chr 11 at 100 kb resolution, colored by log fold change in cfDNA signals. Image: Brown University.
| ||||||||||||||||||||||||||||

