|

CLICK ON weeks 0 - 40 and follow along every 2 weeks of fetal development
|
||||||||||||||||||||||||||||
|
Developmental Biology - Regeneration The Genetics of Regeneration "Only about two percent of the genome makes things like proteins," Andrew Gehrke explains. "We wanted to know: What is the other 98 percent of the genome doing during whole-body regeneration? People have known for some time that many DNA changes cause disease in non-coding DNA regions. But, it is underappreciated as a process, like whole-body regeneration. I think we've only just scratched the surface. We've looked at some switches, but there is another aspect of how the genome interacts on a larger scale, not just how pieces open and close. All of it is important for turning genes on and off; so, I think there are multiple layers to this regulatory nature." Structured Abstract INTRODUCTION Although all animals can heal wounds, some are capable of reconstructing their entire bodies from small fragments of the original organism. Whole-body regeneration requires the interplay of wound signaling, stem cell dynamics, and positional identity, all of which have been investigated at the protein-coding level of the genome. Little is known about how the noncoding portion of the genome responds to wounding to control gene expression and to launch the process of whole-body regeneration. Understanding how these control points (regulatory regions) are activated and then operate during regeneration would uncover how genes connect into networks, ultimately restructuring entire body axes. Networks of transcriptional regulatory genes can reveal important mechanisms for how animals can grow new skin, muscles, or even entire brains. RATIONALE To identify regulatory regions involved in whole-body regeneration, we sequenced the genome of the highly regenerative acoel Hofstenia miamia, commonly known as the three-banded panther worm. Equipped with this genome, we reasoned that applying the assay for transposase-accessible chromatin using sequencing (ATAC-seq) would identify regulatory regions that change in response to amputation and during whole-body regeneration. Further, by analyzing the sequence motifs contained within these regulatory regions, we sought to predict which transcription factors (TFs) control regeneration gene networks. RESULTS The Hofstenia genome assembly totals 950 megabases of sequence, with sufficient contiguity for functional genomics. ATAC-seq data revealed thousands of chromatin regions that respond dynamically during regeneration. A genome-wide scan for TF binding motifs in these regions identified the EGR (early growth response) motif as the most dynamic. By combining RNA interference (RNAi) and RNA-seq, we predicted a set of EGR target genes in Hofstenia. We found that most of these target genes contained EGR binding motifs in neighboring regions of regeneration-responsive chromatin, which failed to respond under egr-RNAi. This functional validation allowed us to build a gene regulatory network (GRN) with EGR as a direct master regulator of downstream regeneration genes. Lastly, by quantifying the binding probabilities of TFs at individual motifs, we identified targets of TFs further downstream of EGR, extending the regeneration GRN. CONCLUSION Using our regulatory data, we inferred a GRN for launching whole-body regeneration in the acoel H. miamia, where the master regulator EGR acts as a putative pioneer factor to directly activate wound-induced genes. This network includes homologs of genes that are involved in regeneration in other species, suggesting that it can serve as a template for direct comparisons of regeneration pathways across distantly related animals. Our approach of combining genome-wide assays for chromatin accessibility with functional studies can be applied to extend the network further in time in Hofstenia regeneration and to construct GRNs for regeneration in other systems. Authors Andrew R. Gehrke, Emily Neverett, Yi-Jyun Luo, Alexander Brandt, Lorenzo Ricci, Ryan E. Hulett, Annika Gompers, J. Graham Ruby, Daniel S. Rokhsar, Peter W. Reddien and Mansi Srivastava1. Acknowledgements This research was supported with funding from the Milton Fund of Harvard University, the Searle Scholars Program, the Smith Family Foundation, the National Science Foundation, the Helen Hay Whitney Foundation, the Human Frontier Science Program, the National Institutes of Health, the Biomedical Big Training Program, UC Berkeley, the Marthella Foskett Brown Chair in Biological Sciences, and the Howard Hughes Medical Institute. Return to top of page | Mar 21 2019 Fetal Timeline Maternal Timeline News 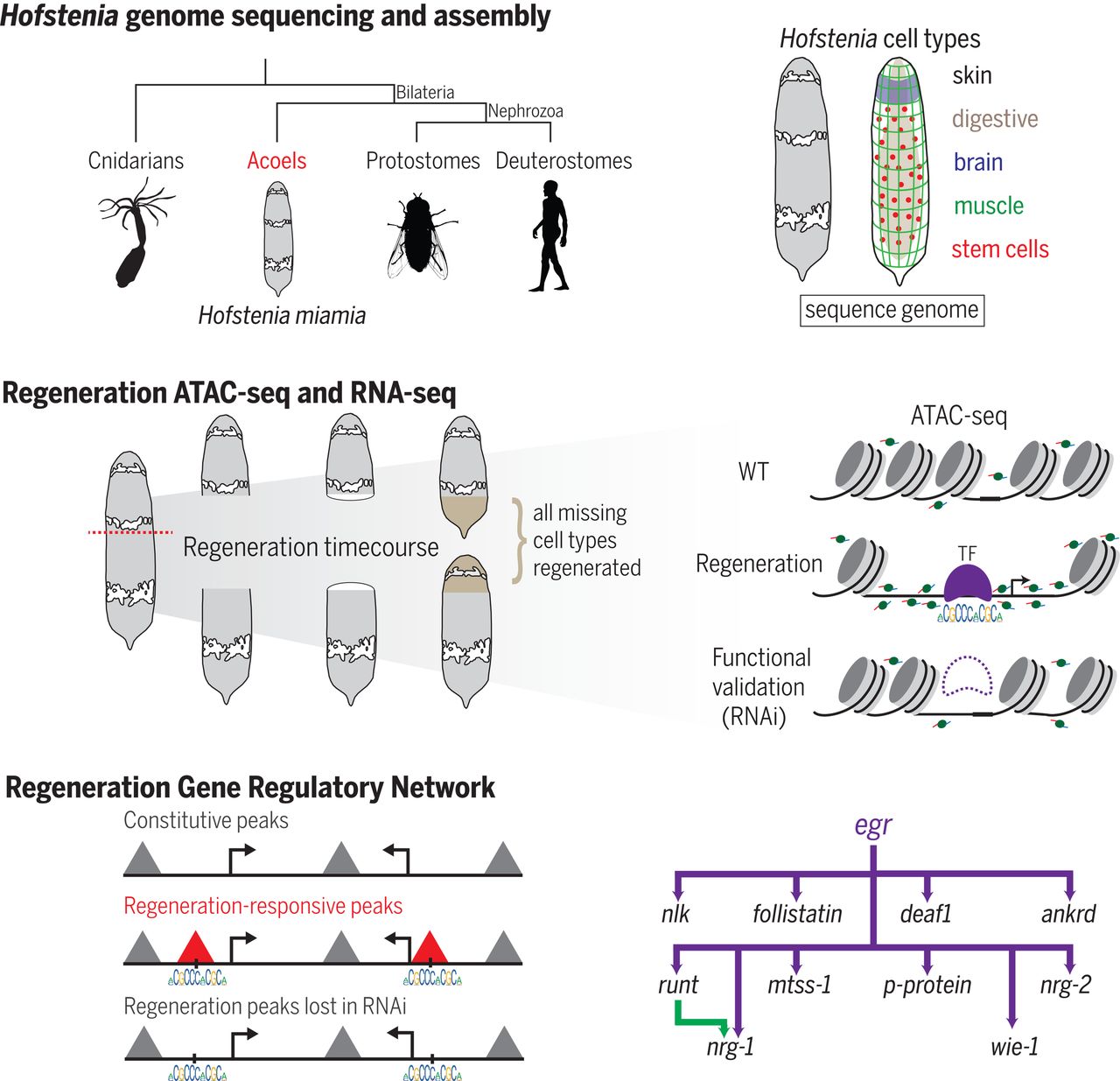 The regulatory landscape of whole-body regeneration. Researchers sequenced the genome and used ATAC-seq to identify thousands of regeneration-responsive regions of chromatin. Combining motif analysis, ATAC-seq, and RNAi, they identified EGR as a master regulator of regeneration in Hofstenia [commonly called the three-banded panther worm] and inferred an EGR-controlled GRN for regeneration. Science: 15 Mar 2019:Vol. 363, Issue 6432.
|
||||||||||||||||||||||||||||

