|
|
Developmental Biology - Stem Cell Specialization
KLF4 Role In Deciding Cell Fate
First event in stem cells becoming organs...
Proteins known as transcription factors - produced from instructions in our DNA - regulate conversion or transcription of DNA into RNA. Regulating transcription is the most common form of gene control.
New research by cell biologists at the University of Toronto (U of T) provides significant insight into the very first decision stem cells go through in order to become specialized cells that make up organs.
Published in Genes & Development the findings implicate a protein's ability to hang around in cells, their stability, as a major factor in controlling whether a stem cell remains a stem cell or transforms into a specialized cell.
Stem cells are regulated by a network of proteins which maintain that cell's ability to become any type of cell, a property known as pluripotency.
These new findings highlight the role of KLF4, one of the transcription factors giving stem cells their unique properties. This discovery was serendipitous as researchers initially set out to investigate how the KLF4 gene is regulated in transcription, turning their attention to the KLF4 protein instead.
"Many previous studies focused on genes that are turned on or off as stem cells are destined to make specific organs. Our work exposes a situation earlier in the process where reducing gene expression by 90 per cent does not affect the amount of protein made. It was a really surprising finding when we first saw the results."
Navroop Dhaliwal PhD, under lab director and professor Jennifer Mitchell PhD, Department of Cell & Systems Biology in the Faculty of Arts & Science at U of T.
Researchers found KLF4 proteins made one day remained functional 24 hours later - a surprise as transcription factors typically only last two or three hours. When they looked at how stem cells differentiate and exit the stem cell state, they found KLF4 becomes unstable during this process — by preventing this breakdown the cells can't differentiate.
Dhaliwal: "We discovered the KLF4 protein is highly stable and locks cells in the stem cell state. These findings have important implications for regenerative medicine as building new organs requires a detailed understanding of how stem cells exit their immature state.
Knowing this, we can now develop more efficient ways to produce patient specific stem cells and differentiate these cells to more mature cells, which will be the focus of my postdoctoral work. Breaking it down, however, released stem cells to specialize and eventually become the different organs of the body."
Dhaliwal and her colleagues believe their findings indicate how KLF4 protein destabilization is a critical step in the ability of a stem cell to become any one of the hundreds of special cell types found in a mature organism.
Beyond its role in stem cells, KLF4 is also involved in numerous cancers. Mechanisms uncovered here may shed light on its role in the development of breast cancer, squamous cell carcinoma and gastrointestinal cancers.
"The data we present highlights the importance of studying both transcriptional control and mechanisms that affect protein abundance. These mechanisms are particularity timely to keep in mind as more and more work shifts to a focus on gene expression using techniques like single cell RNA-sequencing, which would not have revealed the mechanisms we uncovered."
Jennifer Mitchell PhD, Lab Director, Department of Cell & Systems Biology in the Faculty of Arts & Science at U of T.
Abstract
Embryonic stem (ES) cells are regulated by a network of transcription factors that maintain the pluripotent state. Differentiation relies on down-regulation of pluripotency transcription factors disrupting this network. While investigating transcriptional regulation of the pluripotency transcription factor Kruppel-like factor 4 (Klf4), we observed that homozygous deletion of distal enhancers caused a 17-fold decrease in Klf4 transcript but surprisingly decreased protein levels by less than twofold, indicating that posttranscriptional control of KLF4 protein overrides transcriptional control. The lack of sensitivity of KLF4 to transcription is due to high protein stability (half-life >24 h). This stability is context-dependent and is disrupted during differentiation, as evidenced by a shift to a half-life of <2 h. KLF4 protein stability is maintained through interaction with other pluripotency transcription factors (NANOG, SOX2, and STAT3) that together facilitate association of KLF4 with RNA polymerase II. In addition, the KLF4 DNA-binding and transactivation domains are required for optimal KLF4 protein stability. Posttranslational modification of KLF4 destabilizes the protein as cells exit the pluripotent state, and mutations that prevent this destabilization also prevent differentiation. These data indicate that the core pluripotency transcription factors are integrated by posttranslational mechanisms to maintain the pluripotent state and identify mutations that increase KLF4 protein stability while maintaining transcription factor function.
Authors
Navroop K. Dhaliwal, Luis E. Abatti and Jennifer A. Mitchell.
Acknowlegements
Support for the research was provided by the Canadian Institutes of Health Research, the Canada Foundation for Innovation, the Ontario Ministry of Research and Innovation and the Ontario Graduate Scholarship program.
This article is distributed exclusively by Cold Spring Harbor Laboratory Press for the first six months after the full-issue publication date (see http://genesdev.cshlp.org/site/misc/terms.xhtml). After six months, it is available under a Creative Commons License (Attribution-NonCommercial 4.0 International), as described at http://creativecommons.org/licenses/by-nc/4.0/.
Return to top of page.
| |
|
Jun 26 2019 Fetal Timeline Maternal Timeline News
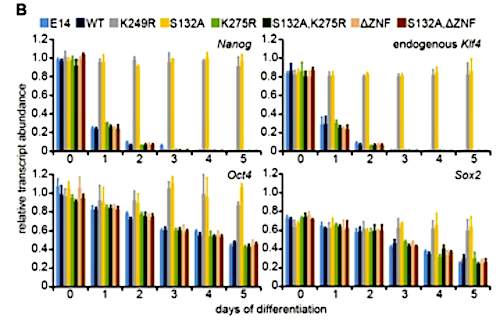 Loss of KLF4 protein stability is required for ES cell differentiation B) Klf4, Nanog, Oct4 and Sox2 were quantified in three biological replications of cells. WT and indicated KLF4-GFP mutant cells reveal only KLF4(S132A)-GFP and KLF4(K249R)-GFP blocked differentiation. Error bars = standard deviation. CREDIT Cold Spring Harbor Laboratory Press
|



