|
|
Developmental Biology - Organ Growth
6 Mammal Gene Networks Profiled
Researchers decode 6 mammal gene networks to find how similar they are...
For the first time, researchers have decoded gene networks that control several major organ systems as they develop before and after birth. Not only in humans, but also in rhesus monkeys, mice, rats, rabbits, and opossums. The molecular biologists at Heidelberg University, Germany analyzed the growth of seven organ systems in each animal: brain, heart, liver, kidney, ovaries, and testicles.
Their work reveales how all of these six gene networks must have originatinated early in the evolution of mammals — more than 200 million years ago in the late Triassic.
A second part of the study focuses on the poorly understood, but largest unit of heredity exchange — RNA genes (ribonucleic acid) — in their paper published in Nature.
Complex interactions by a large number of genes are what turns a fertilized egg into an adult organism. But fitting together all these billions of interactions, necessarily has been restricted to following individual genes and the proteins they create in only a few organ systems. And then, most developmental research is restricted to mice. "The genetic foundations that account for the differences in size, structure, and function of organs in different mammals went largely unknown," explains Prof. Henrik Kaessmann, group leader of the research team at the Center for Molecular Biology of Heidelberg University (ZMBH).
To fully investigate genetic programs in development, Kaessmann's team turned to innovative Next Generation Sequencing technologies (NGS) which enables gene expression analysis of all genes at the same time. Using NGS technology, more than 100 billion gene fragments from proteins and RNA of various mammal organ systems were read all at once. "This allowed us to quantify and compare shifting gene activities over the course of development," explains Margarida Cardoso-Moreira PhD who along with Ioannis Sarropoulos PhD, are the first authors of the two publications.
Bioinformatic analysis performed using high-powered computers at Heidelberg University Computing Center, revealed unique gene activity networks. Networks that function similarly in all mammals studied, including humans.
Researchers also found a surprisingly large number of genes that deviated significantly from one another in various mammal species. These differences must have risen up over the course of evolution, helping to explain specific organ traits in varying species.
For example, Heidelberg researchers were able to identify distinct gene expression patterns controlling human brain development. They also found a surprisingly large number of RNA genes in control of organ development. According to Kaessmann, RNA gene fragments which previously had been difficult to distinguish prior to NGS analysis, now are seen to play a significant role in mammal differentiation.
Additionally, following in the steps of German naturalist Karl Ernst von Baer (1792 - 1876) researchers verified his observation that the earlier in embryo development, the more similar each species is.
In this large-scale study, researchers were now able to genetically confirm with high-level gene pattern differentiation, how each embryo of a species appears molecularly more similar to another the earlier in development it is. Although very similar during these prenatal phases, organ development in all six species studied, increased in complexity as that animal progressed to birth and continued after birth.
"Organ traits that characterise a species do not originate until late in development. Using modern molecular methods, we were able for the first time to confirm a groundbreaking hypothesis in biology stemming from the 19th century."
Henrik Kaessmann PhD, Professor, Molecular Biology, Center for Molecular Biology, Heidelberg University (ZMBH) and group leader of the research team.
Abstract
The evolution of gene expression in mammalian organ development remains largely uncharacterized. Here we report the transcriptomes of seven organs (cerebrum, cerebellum, heart, kidney, liver, ovary and testis) across developmental time points from early organogenesis to adulthood for human, rhesus macaque, mouse, rat, rabbit, opossum and chicken. Comparisons of gene expression patterns identified correspondences of developmental stages across species, and differences in the timing of key events during the development of the gonads. We found that the breadth of gene expression and the extent of purifying selection gradually decrease during development, whereas the amount of positive selection and expression of new genes increase. We identified differences in the temporal trajectories of expression of individual genes across species, with brain tissues showing the smallest percentage of trajectory changes, and the liver and testis showing the largest. Our work provides a resource of developmental transcriptomes of seven organs across seven species, and comparative analyses that characterize the development and evolution of mammalian organs.
Authors
Margarida Cardoso-Moreira, Jean Halbert and Henrik Kaessmann.
Acknowlegements
Researchers from China, Great Britain, Portugal, Russia, Sweden, Switzerland, and the United States contributed to the study. Funding was provided by the European Research Council, the Swiss National Science Foundation, and a Marie Curie grant from the European Union. The data are available in a public access database.
The authors thank R. Arguello, F. Lamanna, I. Sarropoulos, M. Sepp and members of the Kaessmann group for discussion; I. Moreira for developing the Shiny application; M. Sanchez-Delgado and N. Trost for figure assistance; P. Dhakal for assistance collecting rat samples; S. Nef for assistance collecting gonadal samples; the Lausanne Genomics Technology Facility for high-throughput sequencing; the Joint MRC/Wellcome (MR/R006237/1) Human Developmental Biology Resource (HDBR), the Maryland Brain Collection Center and the Chinese Brain Bank Center for providing human samples; and the Suzhou Experimental Animal Center for providing rhesus macaque samples. Computations were performed at the Vital-IT Center of the SIB Swiss Institute of Bioinformatics and at the bwForCluster from the Heidelberg University Computational Center (supported by the state of Baden-Württemberg through bwHPC and the German Research Foundation - INST 35/1134-1 FUGG). M.J.S. was supported by the National Institutes of Health (HD020676); P.J. by the European Research Council (322206, Genewell); Y.E.Z. by the National Natural Science Foundation of China (31771410, 91731302); M.C. by Fundação para a Ciência e Tecnologia through POPH-QREN funds from the European Social Fund and Portuguese MCTES (IF/00283/2014/CP1256/CT0012); S. Anders by the German Research Foundation (SFB 1036). This research was supported by grants from the European Research Council (615253, OntoTransEvol) and Swiss National Science Foundation (146474) to H.K. and by the Marie Curie FP7-PEOPLE-2012-IIF to M.C.-M. (329902).
Return to top of page.
| |
|
Jul 8 2019 Fetal Timeline Maternal Timeline News
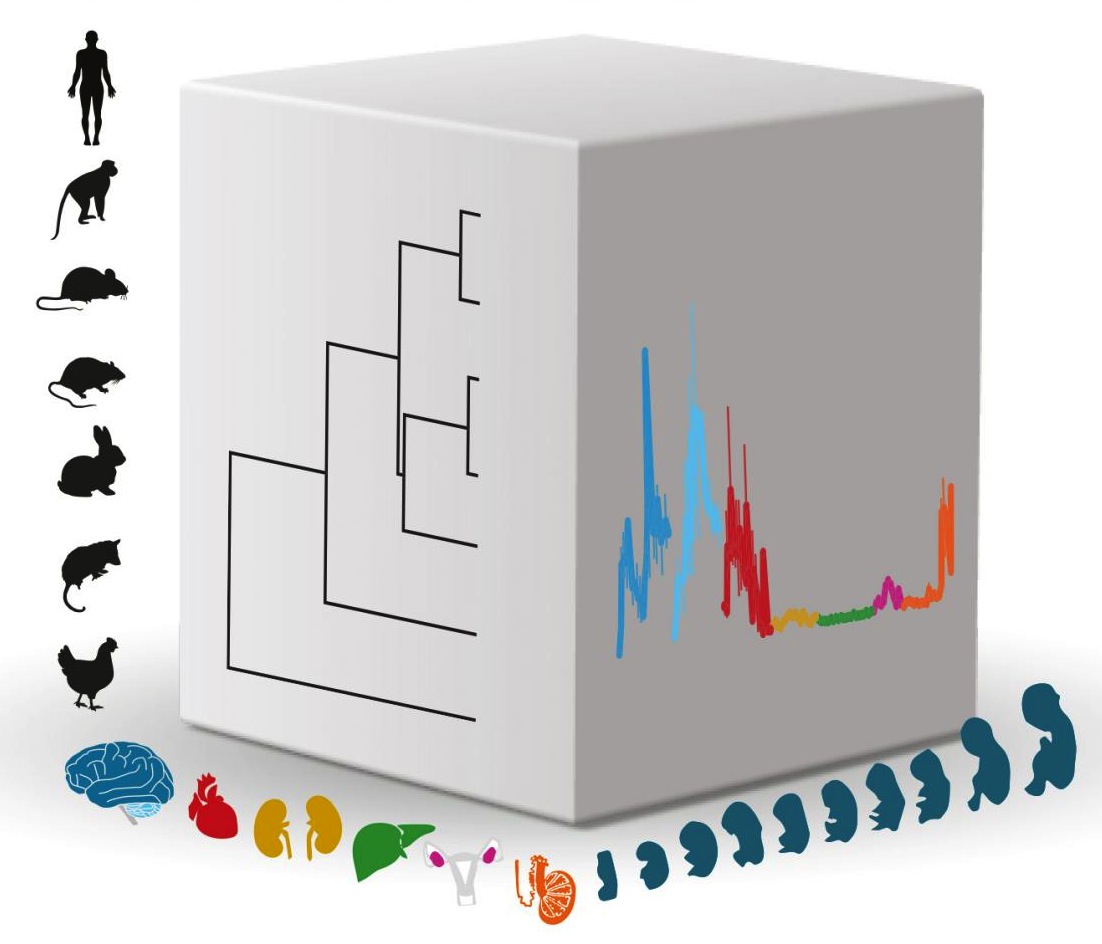 Researchers led by professor Henrik Kaessmann investigated the activity (expression) of genes across mammal organ development in several species. This cube illustrates the three main biological dimensions of their work: 1. species 2. organs and 3. developmental stages. The evolutionary relationships of species are indicated in a tree on the left front of the cube, while the expression of a particular gene as it appears across development in different organs, is indicated on the right front surface. CREDIT Kaessmann research group
|



