|
|
Developmental Biology - Cell Imaging
Frankenbodies
Not an antibody, but a frankenbody. A new tool for live-cell imaging...
Antibodies are the biomolecules our immune systems deploy to find, tag and destroy invading pathogens. They work by binding to specific targets, called epitopes, on the surfaces of antigens - like locks to keys.
For many decades, scientists cleverly exploited the selective tagging mechanism of natural antibodies to engineer antibody-based probes, allowing scientists to purify and study different types of proteins within cells. One tried and true technique, epitope tagging, involves fusing an epitope to a protein of interest and using fluorescently labeled antibodies to make those proteins visible. However, it only works on fixed, dead, cells.
Now, a cross-disciplinary team of researchers from Colorado State University (CSU) and the Tokyo Institute of Technology have added a new tool to the arsenal of antibody-based probes. One with a powerful distinction — this genetically encoded probe works in living cells. The work, led by CSU Monfort Professor Tim Stasevich and Tokyo Tech Professor Hiroshi Kimura, is described July 3 in the journal Nature Communications.
According to first author Ning Zhao, a postdoctoral researcher in Stasevich's lab who designed most of the experiments, their new probe is affectionately called a "frankenbody."
Like stitching new limbs onto a body, the scientists have taken the binding regions of a normal antibody, its "sticky parts," and grafted them to a scaffold that remains stable in live cells and retains the specificity of the antibody.
"We're interested in intracellular antibodies because you can use them as imaging reagents in a live cell. You don't need a tag, like a Green Fluorescent Protein, instead you have this fluorescent antibody that will bind to the protein you want to visualize."
Timothy Stasevich PhD, Assistant Professor, Department of Biochemistry and Molecular Biology, Colorado State University, USA.
The new probe will be a useful complement to the green fluorescent protein (GFP). GFP is a widespread biochemistry tool and subject of a Nobel Prize. It involves genetically fusing a light-up green tag to a protein of interest. However, the GFP is limited by its relatively large size and the time it takes to fluoresce; with the CSU researchers' new probe, the tag is smaller and becomes fluorescent faster, so the "birth" of a protein of interest can be captured in real time.
With the goal of making their tool immediately useful, the scientists designed their probe to work with the classic HA tag. HA is a widely used small linear epitope tag that's derived from a portion of the human influenza virus protein hemagglutinin.
"For the longest time, people have been looking at HA-tagged proteins in fixed, dead cells," Stasevich adds. "Now we can image the dynamics of those proteins in live cells."
The possibilities of how scientists may use the new probe are limitless. Stasevich's lab is particularly interested in studying RNA translation, and they plan to use their new system to more easily design new RNA imaging experiments.
Added Zhao, the HA tag is tiny - a chain of just nine amino acids - and the probe is genetically encoded on a plasmid that can be easily transferred into a cell. This is in contrast to traditional antibodies, which can cost a lab several hundreds of dollars per order, suffer from lot-to-lot variability, and are difficult to get into cells. The new probe from Stasevich's team therefore provides a low-cost solution for protein and RNA imaging.
In the paper, the scientists demonstrated some applications, including single-protein tracking, single-RNA translation imaging, and amplified fluorescence imaging in zebrafish embryos. All of these experiments are more challenging when using traditional fluorescent protein tags.
"We have several new imaging reagents in the works that build off of this success, so I see great things ahead."
Timothy Stasevich PhD
Abstract
To expand the toolbox of imaging in living cells, we have engineered a single-chain variable fragment binding the linear HA epitope with high affinity and specificity in vivo. The resulting probe, called the HA frankenbody, can light up in multiple colors HA-tagged nuclear, cytoplasmic, membrane, and mitochondrial proteins in diverse cell types. The HA frankenbody also enables state-of-the-art single-molecule experiments in living cells, which we demonstrate by tracking single HA-tagged histones in U2OS cells and single mRNA translation dynamics in both U2OS cells and neurons. Together with the SunTag, we also track two mRNA species simultaneously to demonstrate comparative single-molecule studies of translation can now be done with genetically encoded tools alone. Finally, we use the HA frankenbody to precisely quantify the expression of HA-tagged proteins in developing zebrafish embryos. The versatility of the HA frankenbody makes it a powerful tool for imaging protein dynamics in vivo.
Authors
Ning Zhao, Kouta Kamijo, Philip D. Fox, Haruka Oda, Tatsuya Morisaki, Yuko Sato, Hiroshi Kimura and Timothy J. Stasevich.
Acknowledgements
The authors thank all members of the Stasevich lab for input and helpful suggestions, especially Kenneth Lyon for removing the HA epitopes from the Sun-GFP construct (Addgene #60907) and Lara Perinet for helping with plasmid preparation. We also thank members of the CSU TagTeam; in particular, Dr. Chris Snow, Dr. Brian Geiss, and Steve Foster, for valuable discussions. Finally, we thank Laurie S. Minamide, Dr. James R. Bamburg and Dr. Michael M. Tamkun of CSU for supplying neurons, Dr. Michael M. Tamkun of CSU for supplying pCMV-mRuby-Kv2.1 plasmid and Dr. Luke Lavis of Janelia for supplying JF646 dye in this study. The research reported in this publication was supported by Colorado State University’s Office of the Vice President for Research Catalyst for Innovative Partnerships Program. The content is solely the responsibility of the authors and does not necessarily represent the official views of the Office of the Vice President for Research. This work was also funded through an award to T.J.S. by the NIH (R35GM119728). T.J.S. is also supported by funds from the Boettcher Foundation’s Webb-Waring Biomedical Research Program. H.K. was supported by KAKENHI JP18H05527.
The team of researchers is grateful for the support they received from the National Institutes of Health (grant No. R35GM119728); the Boettcher Foundation's Webb-Waring Biomedical Research Program; the Japan Society for the Promotion of Science (KAKENHI grant No. JP18H05527); and from CSU's Catalyst for Innovative Partnerships program and Tokyo Tech's World Research Hub Initiative. The work strengthens ties between CSU and the Tokyo Institute of Technology.
Return to top of page.
| |
|
Jul 9 2019 Fetal Timeline Maternal Timeline News
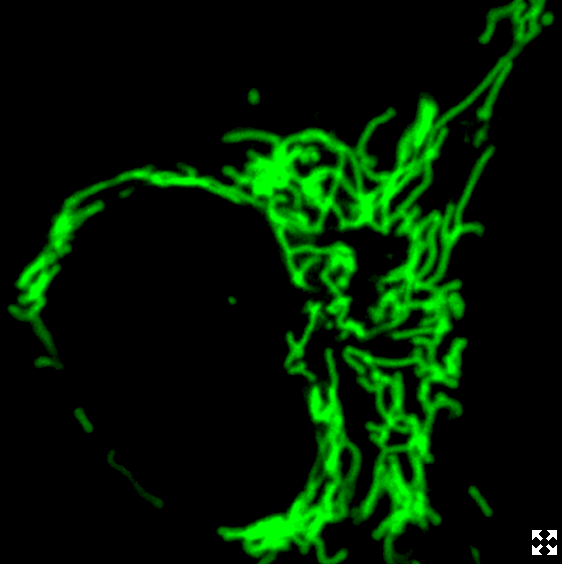 This is a Frankenbody labeling mitochondria. It works in living cells by binding to regions of a normal antibody, its "sticky parts," and grafting them to a scaffold. The scaffold remains stable in live cells while retaining the specificity of the antibody. CREDIT Ning Zhao/Colorado State University.
|



