|
|
Developmental Biology - RNA
How RNA Molecules Fold
In a first for cell biology, scientists observe ribosome assembly in real time...
An imaging feat clarifies how RNA molecules fold - and might one day translate into better medicines for a host of diseases.
Scientists from Scripps Research and Stanford University has recorded in real time a key step in the assembly of ribosomes - the complex and evolutionarily ancient "molecular machines" that make proteins in cells and are essential for all life forms.
The achievement, reported in Cell, reveals in unprecedented detail how strands of ribonucleic acid (RNA), cellular molecules that are inherently sticky and prone to misfold, are "chaperoned" by ribosomal proteins into folding properly and forming one of the main components of ribosomes.
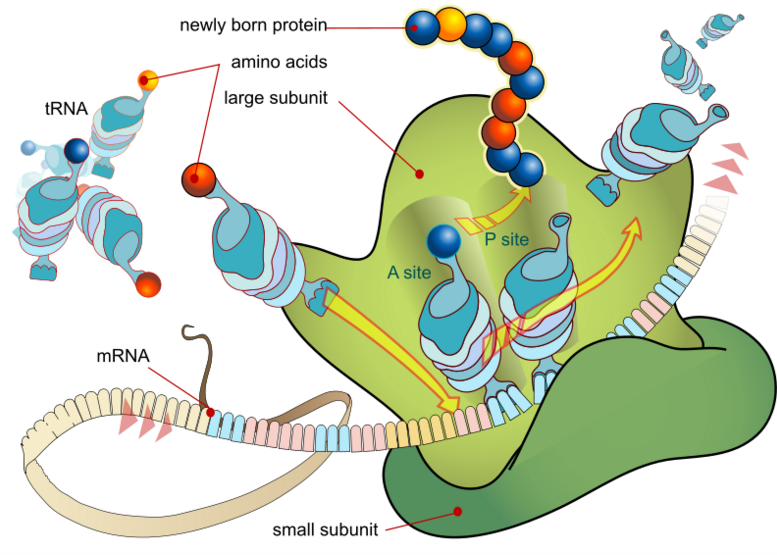 Ribosomes are the translational apparatus of a cell.
Ribosomes are the translational apparatus of a cell.
GREEN = Ribosome. Wikipedia
"In contrast to what had been the dominant theory in the field, we revealed a far more chaotic process. It's not a sleek Detroit assembly line - it's more like a trading pit on Wall Street."
James R. Williamson PhD, Professor, Department of Integrative Structural & Computational Biology, The Scripps Research Institute, La Jolla, California, USA.
For the study, Williamson's lab collaborated with the lab of Joseph Puglisi PhD, professor, department of Structural Biology, Stanford University School of Medicine, California. Their work is a significant feat of basic cell biology and should enable important advances in medicine. For example, some current antibiotics work by inhibiting bacterial ribosomes — their new research opens up the possibility of designing future antibiotics that target bacterial ribosomes with greater specificity - thus fewer side effects.
"We now can examine in detail how RNAs fold while they are being synthesized and proteins are assembling on them ... a very difficult thing to study in biology because it involves several distinct biological processes dependent on each other having to be detected simultaneously."
Olivier Duss PhD, Postdoctoral Research Fellow, Department of Integrative Structural & Computational Biology, Scripps Research.
The team used an advanced imaging technology called "zero-mode waveguide single-molecule fluorescence microscopy," adapted in recent years for real-time tracking of RNAs and proteins. Ribosomes are both RNA and proteins, a molecular partnership going back nearly to the dawn of life on Earth.
Their proof-of-principle study published in Nature Communications last year used this approach to record an early, brief and relatively well-studied stage of the bacterium E. coli ribosome assembly: the transcription/copying from its corresponding gene - a ribosomal RNA - and it's initial interactions with a ribosomal protein.
In today's 2019 study, the team extended their approach by tracking not only transcription of a ribosomal RNA, but also its real-time folding. This provided a detailed look at a complex, mysterious part of E. coli ribosome assembly — the formation of an entire major component/domain of an E. coli ribosome, plus its eight protein partners incorporated into the same structure.
A key finding is that ribosomal proteins guide RNA folding through multiple temporary interactions long before nestling into a final RNA-protein molecule. Researchers believe this hints at the existence of unknown RNA assembly factors, most likely proteins, not present in their lab-dish-type imaging experiments, but within cells to boost the efficiency of RNA folding.
"Our study indicates that within ribosomal RNA-folding, and perhaps generally in RNA-folding in cells, many proteins help fold RNA through weak, transient and semi-specific interactions."
Olivier Duss PhD.
The team will now be able to extend this research further to study not only the rest of ribosome assembly, involving multiple RNA strands and dozens of proteins, but also the many other types of RNA-folding and RNA-protein interactions within cells. In principle, this will produce insights into how RNAs misfold and how these events could be corrected. Scientists believe many diseases involve, or potentially involve, improper folding and related processing of cell RNAs.
Treatments that already target ribosomes might also be improved. Some current antibiotics, including a class known as aminoglycosides, work by binding to sites on bacterial ribosomes not present on human ribosomes. These drugs can have side effects as they also impair ribosomes of good bacteria like those in our gut.
"When we understand more fully how bacterial ribosomes assemble and function, we could potentially target them in ways that affect a narrower group of harmful bacterial species and spare the good ones — reducing side effects for patients," Duss explains.
"Because ribosomes function as protein makers, they are crucial to the survival of fast-growing tumor cells. Several classes of cancer drug already work by slowing ribosome formation in one way or another. A better understanding of human ribosomes would, in principle, enable its assembly to be targeted more precisely and effectively in blocking cancer growth."
Olivier Duss PhD.
Highlights
• Real-time tracking of transcription, nascent RNA folding, and protein binding
• Nascent RNA folding is complicated by native long-range RNA-RNA interactions
• Late-binding ribosomal proteins chaperone nascent rRNA folding early in assembly
• Protein-RNA binding dynamics cooperatively decrease during ribosome assembly
Summarybr>
Ribosome assembly is an efficient but complex and heterogeneous process during which ribosomal proteins assemble on the nascent rRNA during transcription. Understanding how the interplay between nascent RNA folding and protein binding determines the fate of transcripts remains a major challenge. Here, using single-molecule fluorescence microscopy, we follow assembly of the entire 3? domain of the bacterial small ribosomal subunit in real time. We find that co-transcriptional rRNA folding is complicated by the formation of long-range RNA interactions and that r-proteins self-chaperone the rRNA folding process prior to stable incorporation into a ribonucleoprotein (RNP) complex. Assembly is initiated by transient rather than stable protein binding, and the protein-RNA binding dynamics gradually decrease during assembly. This work questions the paradigm of strictly sequential and cooperative ribosome assembly and suggests that transient binding of RNA binding proteins to cellular RNAs could provide a general mechanism to shape nascent RNA folding during RNP assembly.
Authors
Olivier Duss, Galina A. Stepanyuk, Joseph D. Puglisi, James R. Williamson.
Acknowledgments
The other co-author of the study, "Transient Protein-RNA Interactions Guide Nascent Ribosomal RNA Folding," was Galina Stepanyuk, PhD, of Scripps Research.
Support for the research was provided by the National Institutes of Health (R01 GM051266, R01 GM113078, and R01 GM053757), the Swiss National Science Foundation (P2EZP3-152131, P300PA-160978), and the Human Frontier Science Program (LT000628/2015-L).
Return to top of page.
| |
|
Nov 27 2019 Fetal Timeline Maternal Timeline News

One day, better medicines might be designed to regulate how RNA
molecules fold affecting a host of diseases. CREDIT Scripps Research.
|




