|
|
Developmental Biology - Chromatin Structure
Chromatin Folds Into The Cell Nucleus Like A 3D Forest
Chromatin helps pack about six feet of DNA into each cell nucleus in our body...
A single human cell contains all the genetic instructions needed to make an entire person. This gene information is managed and processed by complex machinery called chromatin which is a mix of DNA and proteins that exists inside chromosomes. Its function and role in disease is increasingly of interest to science.
A Northwestern University research team - using mathematical modeling and optical imaging techniques they developed - has discovered how chromatin folds at the single-cell level.
 The researchers found chromatin folds into a variety of tree-like domains along a chromatin backbone. Both small and large areas exist in this construction, much as a mixed forest of trees grows from the forest floor. The overall structure appears to be a microscale forest in 3D.
The researchers found chromatin folds into a variety of tree-like domains along a chromatin backbone. Both small and large areas exist in this construction, much as a mixed forest of trees grows from the forest floor. The overall structure appears to be a microscale forest in 3D.
Chromatin is responsible for packing DNA into the cell nucleus. In humans, that's about six feet of DNA per cell. This new work suggests chromatin is more structured and hierarchical in single cells than previously thought. Learning how chromatin operates will help scientists understand what is going wrong with chromatin in cancers and other diseases.
"By integrating theoretical and experimental work, we produced a new chromatin folding picture that helps us see how the 3D genome is organized at the single-cell level," explains Igal Szleifer, the Christina Enroth-Cugell Professor of Biomedical Engineering at Northwestern's McCormick School of Engineering. Szleife co-led the research team with Vadim Backman.
Details of the interdisciplinary study published Jan. 10, 2020 in the journal Science Advances.
"If genes are the hardware, chromatin is the software. If the structure of chromatin changes, it can alter the processing of information stored in the genome — but it does not alter the genes themselves. Understanding chromatin folding holds a key to understanding how cells differentiate and how cancer happens."
Vadim Backman PhD, Walter Dill Scott Professor of Biomedical Engineering; Director, Center for Physical Genomics and Engineering; Professor of Medicine, Biochemistry and Molecular Genetics, the Feinberg School of Medicine; Program Leader, Cancer and Physical Sciences, the Robert H. Lurie Comprehensive Cancer Center, Northwestern University; member Chemistry of Life Processes Institute.
Advances in genomic, imaging and information technologies are just beginning to enable scientists to better understand how chromatin works. Northwestern researchers used a Partial Wave Spectroscopic (PWS) microscope, optical imaging developed by Backman and colleagues, to look deep into live cells and "sense" alterations in chromatin packing. They also used electron imaging.
"Our paradigm-shifting picture of chromatin folding is an important missing piece in the holistic view of genomic structure," adds Kai Huang, the study's first author and postdoctoral fellow in Backman's research group. "The results should inspire new strategies to fight cancer."
Abstract
With the textbook view of chromatin folding based on the 30-nm fiber being challenged, it has been proposed that interphase DNA has an irregular 10-nm nucleosome polymer structure whose folding philosophy is unknown. Nevertheless, experimental advances suggest that this irregular packing is associated with many nontrivial physical properties that are puzzling from a polymer physics point of view. Here, we show that the reconciliation of these exotic properties necessitates modularizing three-dimensional genome into tree data structures on top of, and in striking contrast to, the linear topology of DNA double helix. These functional modules need to be connected and isolated by an open backbone that results in porous and heterogeneous packing in a quasi–self-similar manner, as revealed by our electron and optical imaging. Our multiscale theoretical and experimental results suggest the existence of higher-order universal folding principles for a disordered chromatin fiber to avoid entanglement and fulfill its biological functions..
Authors
Kai Huang, Yue Li, Anne R. Shim, Ranya K. A. Virk, Vasundhara Agrawal, Adam Eshein, Rikkert J. Nap, Luay M. Almassalha, Vadim Backman and Igal Szleifer.
Acknowledgments
The authors thank members of the Szleifer and Backman laboratories for their insights and comments. We thank Y. Q. Gao for useful discussion. Funding: We acknowledge funding from the NSF (Biol & Envir Inter of Nano Mat 1833214, EFRI research project 1830961) and the NIH (National Cancer Institute R01 CA228272 and R01 CA225002). This research was supported, in part, through the computational resources and staff contributions provided by the Genomics Compute Cluster, which is jointly supported by the Feinberg School of Medicine, the Center for Genetic Medicine, and Feinberg’s Department of Biochemistry and Molecular Genetics, the Office of the Provost, the Office for Research, and Northwestern Information Technology. The Genomics Compute Cluster is part of Quest, Northwestern University’s high-performance computing facility, with the purpose to advance research in genomics. Author contributions: K.H., V.B., and I.S. conceived the project. K.H. developed the theory and model. Y.L. performed the ChromSTEM experiments and analyzed the data. A.R.S. and R.J.N. participated in the modeling. V.A. and A.E. performed PWS experiments and analyzed the data. R.K.A.V. analyzed the publicly available Hi-C data on heat shock. K.H. wrote the original draft, and all authors reviewed and edited the manuscript. V.B. and I.S. secured the funding and supervised the project. Competing interests: The authors declare that they have no competing interests. Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. Additional data related to this paper may be requested from the authors.
Return to top of page.
| |
|
Jan 15 2020 Fetal Timeline Maternal Timeline News
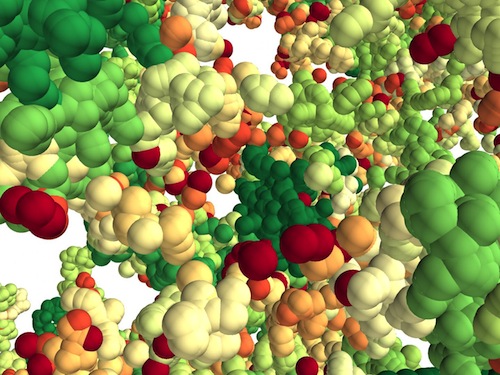 A 3D forest of chromatin predicted by the researchers' model. Tree domains are colored from red to green according to their sizes. Each particle represents 2 kilobases of DNA. CREDIT
Northwestern University.
|




