|
|
Developmental Biology - ALS/Lou Gehrig's disease
Linking ALS Genes To Enzyme - Gemin3
A new study holds promise for treating and perhaps slowing down ALS/Lou Gehrig's disease...
The enzyme Gemin3 is now identified as the molecular 'bridge' between genes whose mutation or disruption causes amyotrophic lateral sclerosis (ALS)/Lou Gehrig's disease - the shrinking of muscle neurons and those muscles they serve. This information comes to us from a new study in Nature Scientific Reports by scientists at the University of Malta.
ALS robs patients of their ability to walk, eat or breath. This late-onset neurodegenerative disease destroys motor neurons — the long nerve cells in the brain and spinal cord that signal muscles to respond. Signals from these nerves gradually shrink away from muscles, which weaken and die.
There is no cure, eventually the disease is fatal.
Genetics contributes significantly to the development of ALS. Mutations in any of an ever-growing list of genes have been identified to cause ALS with TDP-43, FUS and SOD1 featured at the top. Together these 3 genes are responsible for a large percentage of ALS cases with a family history.
"We have been perplexed by the seemingly diverse functions of genes linked to ALS. The lack of commonality complicates the process for developing treatments that are broadly beneficial," explains study leader and researcher Dr Ruben J. Cauchi PhD. Cauchi is a senior lecturer at the University of Malta's Faculty of Medicine & Surgery, and principal investigator at the University of Malta's Centre for Molecular Medicine and Biobanking.
Through research on fruit flies, Cauchi's lab was able to identify a gene called Gemin3 that when even mildly disturbed triggers worsening of ALS symptoms caused by genes TDP-43, FUS or SOD1. Gemin3 produces an enzyme which may be possible to re-tune to alter its effect.
"Our findings point to an overlap in disease-causing mechanisms underlying each different ALS-causing gene. This can potentially unveil new targets for therapies that are effective in a wide range of ALS patients," adds Dr Cauchi.
Gemin3 has long been known for its alliance with the survival motor neuron (SMN) protein. A deficiency of SMN causes spinal muscular atrophy (SMA), a motor neuron disease that strikes infants.
Gemin3's activity is crucial for building the splicing machinery which edits a cell's genetic instructions. Earlier discoveries by this research group also linked Gemin3 to several key players in this delicate process.
Right now, the research team is determining whether targeting multiple players in the pathway uncovered by Gemin3 can improve neuron response with ALS. The results of their efforts may potentially pave the way for effective treatments for a broad group of ALS patients.
Abtract
The predominant motor neuron disease in infants and adults is spinal muscular atrophy (SMA) and amyotrophic lateral sclerosis (ALS), respectively. SMA is caused by insufficient levels of the Survival Motor Neuron (SMN) protein, which operates as part of the multiprotein SMN complex that includes the DEAD-box RNA helicase Gemin3/DDX20/DP103. C9orf72, SOD1, TDP-43 and FUS are ranked as the four major genes causing familial ALS. Accumulating evidence has revealed a surprising molecular overlap between SMA and ALS. Here, we ask the question of whether Drosophila can also be exploited to study shared pathogenic pathways. Focusing on motor behaviour, muscle mass and survival, we show that disruption of either TBPH/TDP-43 or Caz/FUS enhance defects associated with Gemin3 loss-of-function. Gemin3-associated neuromuscular junction overgrowth was however suppressed. Sod1 depletion had a modifying effect in late adulthood. We also show that Gem3ΔN, a helicase domain deletion mutant, retains the ability to interact with its wild-type counterpart. Importantly, mutant: wild-type dimers are favoured more than wild-type: wild-type dimers. In addition to reinforcing the link between SMA and ALS, further exploration of mechanistic overlaps is now possible in a genetically tractable model organism. Notably, Gemin3 can be elevated to a candidate for modifying motor neuron degeneration.
Authors
Rebecca Cacciottolo, Joanna Ciantar, Maia Lanfranco, Rebecca M. Borg, Neville Vassallo, Rémy Bordonné and Ruben J. Cauchi.
Acknowledgments
The authors are indebted to Matthew Camilleri for unwavering technical and administrative support. Thanks also goes to Julia Camilleri for assistance with experiments. We are also very grateful to Aaron Voigt, Magalie Lecourtois, Jane Wu, Brian McCabe, and Fabian Feiguin for fly stocks. This work was supported by the University of Malta Research Fund to RJC, and the Malta Council for Science & Technology Internationalisation Partnership Award to RJC. RC was supported by the Erasmus+ programme of the EU. ML was supported by an Endeavour Scholarship (Malta), part-financed by the EU - European Social Fund under Operational Programme II - Cohesion Policy 2014–2020, "Investing in human capital to create more opportunities and promote the well-being of society." RMB was supported by a Bjorn Formosa Scholarship for Advanced Research into ALS/MND funded by the non-profit organisation, ALS Malta Foundation, facilitated by the Research Trust (RIDT) of the University of Malta.
Study co-authors are Rebecca Cacciottolo, Joanna Ciantar, Maia Lanfranco, Rebecca Borg and Prof Neville Vassallo from the University of Malta, and Dr Remy Bordonne from the Institut de Genetique Moleculaire de Montpellier (CNRS/Universite de Montpellier). Research in Dr. Cauchi's laboratory is founded by the Malta Council for Science & Technology, an Endeavour Scholarship (part-financed by the European Social Fund), ALS Malta Foundation and the University of Malta's Research Trust (RIDT).
Return to top of page.
| |
|
Jan 24 2020 Fetal Timeline Maternal Timeline News
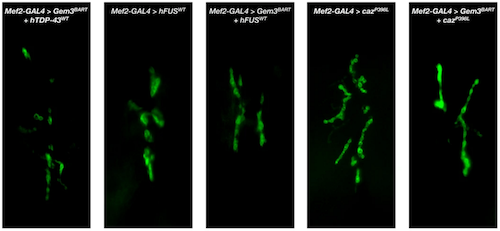 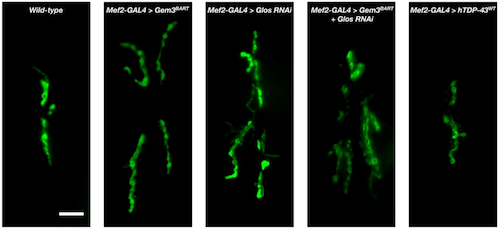 Images of how neuromuscular junctions (NMJ)s stimulate front longitudinal muscles in fly larvae (scale bar = 10µm). Visual inspection reveals how compared to normal (wild-type) flies, muscle-directed function of Gem3BART induced overgrowth of neuromuscular junctions (NMJ) restored by over expression of genes: hTDP-43WT, hFUSWT and to a lesser degree, cazP398L. An overgrowth phenotype is also obvious with reduced Glos, or in flies expressing cazP398L. Observations were confirmed upon numbers of NMJ areas. CREDIT University of Malta Research Trust (RIDT).
|




