|
|
Developmental Biology - CRISPR One Letter Editing
CRISPR Slows ALS Progression In Mice
New CRISPER editing technique changes one letter of DNA sequence without cutting through both DNA strands...
Using a new CRISPR gene-editing technique, scientists from the University of Illinois at Urbana-Champaign inactivated a gene responsible for an inherited form of amyotrophic lateral sclerosis (ALS) - a debilitating and fatal neurological disease with no cure.
The novel treatment slowed an aggressive form of ALS disease in mice, improving muscle function and extending mouse lifespan.
According to bioengineering professor Thomas Gaj PhD, who co-led the study with bioengineering professor Pablo Perez-Pinera: "ALS unfortunately has few treatment options. This is an important first step in showing that this new form of gene editing could be used to potentially treat the disease."
The method relies on an emerging gene-editing technology known as CRISPR base editors.
Traditional CRISPR gene-editing technologies cut both strands of a DNA molecule, which can introduce a variety of errors in the DNA sequence, limiting its efficiency and potentially leading to a number of unintended mutations in the genome. The Illinois group instead used base editing, which changed just one letter of the DNA sequence into another without cutting through both DNA strands.
Gaj explains: "Base editors are too large to be delivered into cells, using one of the most promising and successful gene therapy vectors known — an Adeno-Associated Virus," called an AAV.
In 2019, Perez-Pinera's group developed a method of splitting the base editor proteins in half to be delivered by two separate AAV particles. Once inside the cell, these 2 halves reassembled into a full-length base editor protein.
Combining the power of AAV gene delivery and split-base editors, Gaj and Perez-Pinera targeted and permanently disabled a mutant SOD1 gene responsible for @ 20% of inherited forms of ALS.
These results are published in the journal Molecular Therapy.
"Many ALS studies are focused on preventing or delaying the onset of the disease. However, in the real world, most patients are not diagnosed until symptoms are advanced," said graduate student Colin Lim. "Slowing progression, rather than preventing it, may have a greater impact on patients." Lim is the co-first author of the study along with graduate students Michael Gapinske and Alexandra Brooks.
Researchers first tested the SOD1 base editor in human cells to verify reassembly of the split CRISPR base editor and inactivation of the SOD1 gene. They then injected AAV particles encoding the base editors into spinal columns of mice carrying a mutant SOD1 gene — which causes a particularly severe form of ALS paralyzing mice within a few months following birth.
The disease progressed more slowly in treated mice, which had improved motor function, greater muscle strength and less weight loss. Researchers saw an 85% increase in time between late stage onset and end stage of the disease, as well as increased overall survival.
"We were excited to find that many of the improvements happened well after the onset of the disease. This told us that we were slowing the progression of the disorder," said Gapinske.
The base editor introduces a stop signal near the start of the SOD1 gene, so it stops the cell from making malfunctioning protein no matter which genetic mutation a patient has. However, it potentially disrupts the healthy version of the gene, so researchers are exploring ways to target only the gene's mutant copy in future research.
Gaj: "Moving forward, we are thinking about how we can bring this and other gene-editing technologies to the clinic so that we can someday treat ALS in patients. For that, we have to develop new strategies capable of targeting all cells involved in the disease. We also have to evaluate the efficiency and safety of this approach in other clinically relevant models."
The split base editor approach has potential for treating other diseases with a genetic basis as well, adds Perez-Pinera.
Though ALS was the first demonstration of the tool, studies are underway applying it to Duchenne Muscular Dystrophy and Spinal Muscular Atrophy.
Abstract
Amyotrophic lateral sclerosis (ALS) is a debilitating and fatal disorder that can be caused by mutations in the superoxide dismutase 1 (SOD1) gene. Although ALS is currently incurable, CRISPR base editors hold the potential to treat the disease through their ability to create nonsense mutations that can permanently disable the expression of the mutant SOD1 gene. However, the restrictive carrying capacity of adeno-associated virus (AAV) vectors has limited their therapeutic application. In this study, we establish an intein-mediated trans-splicing system that enables in vivo delivery of cytidine base editors (CBEs) consisting of the widely used Cas9 protein from Streptococcus pyogenes. We show that intrathecal injection of dual AAV particles encoding a split-intein CBE engineered to trans-splice and introduce a nonsense-coding substitution into a mutant SOD1 gene prolonged survival and markedly slowed the progression of disease in the G93A-SOD1 mouse model of ALS. Adult animals treated by this split-intein CRISPR base editor had a reduced rate of muscle atrophy, decreased muscle denervation, improved neuromuscular function, and up to 40% fewer SOD1 immunoreactive inclusions at end-stage mice compared to control mice. This work expands the capabilities of single-base editors and demonstrates their potential for gene therapy.
Authors
Colin K.W. Lim, Michael Gapinske, Alexandra K. Brooks, Wendy S. Woods, Jackson E. Powell, M. Alejandra Zeballos C., Jackson Winter, Pablo Perez-Pinera and Thomas Gaj.
Acknowledgements
The Muscular Dystrophy Association, the Judith and Jean Pape Adams Foundation, the American Heart Association and the National Institutes of Health supported this work.
Dr Adashi serves as cochair of the Safety Advisory Board of Ohana Biosciences, Inc.
Return to top of page.
| |
|
Feb 27 2020 Fetal Timeline Maternal Timeline News
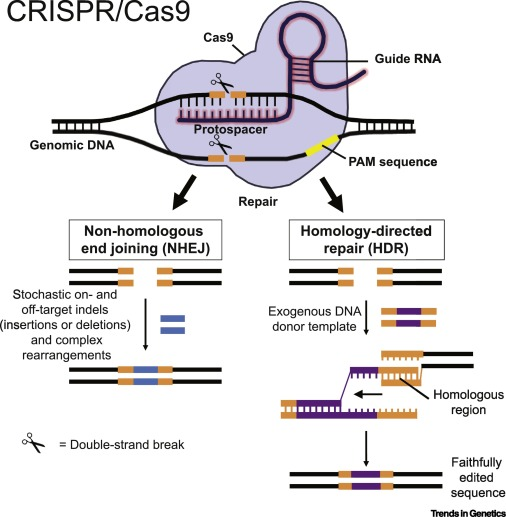 CRISPR/Cas9-Mediated Genome Editing.
The RNA and CRISPR/Cas9 complex first locates the gene region to be edited by identifying adjacent matching protospacer sequences — (PAM, yellow line) on opposite strands. The DNA is then cut producing double-strand DNA breaks (DSBs). Non-homologous end joining (NHEJ) and homology-directed repair (HDR) occurs with two repair mechanisms — leading to error-prone insertions. CREDIT University of Illinois at Urbana-Champaign
|



