|
|
Developmental Biology - Ancient Proteins Inform Modern Cell Division
Discovering The Origin of Modern Proteins
By bringing the ancient history of two proteins back to life, researchers catch a glimpse of how a billion year old molecular partnership works...
Biochemist Dorothee Kern and her colleagues reconstructed two extinct forms of cell division proteins. They then watched as one of those proteins boosted the activity of the other. What Kern's team observed represents the earliest known moment when two ancient proteins interacted.
They reported their finding February 21 in the journal Science.
That interaction, called allosteric regulation or AR, now only occurs between modern proteins and is an indispensable tool in cells. Yet how it evolved was unknown. As cells developed more elaborate networks of protein activity grew in response to evolutionary forces. In order to survive, cells had to create more nuanced control of protein production. AR represents when one proteinbinds to an enzyme and induces that enzyme into an inactive form, explains Kern.
"Allosteric regulation [AR] is arguably the biggest evolutionary step in the development of higher organisms."
Dorothee Kern PhD, Department of Biochemistry and Howard Hughes Medical Institute, Brandeis University, Waltham, Massachusetts, USA.
Kern wants to unravel how proteins, along with other molecules. work together at the atomic level as well as in 'real' time. Pinpointing when proteins send out cell signals to bind to another molecule and kickstart chemical reactions — might eventually give scientists a new window into drug creation adhering to these now obscure rules.
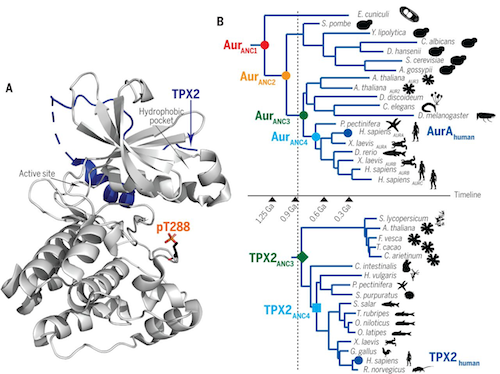
She chose to study a protein called Aurora A as it is vital to cell division and abnormalities in cell division can drive the growth of cancers. Also because Aurora A is turned on by a well known protein partner TPX2.
During cell division, Aurora A helps distribute chromosomes equally between daughter cells:
• TPX2 binds to an "allosteric site" on Aurora A.
• next TPX2 stimulates Aurora A to the point of action, switching the protein into high gear.
Kern wanted to find out how these two proteins' partnership evolved, knowing that organisms throughout the evolutionary tree make some version of both Aurora A and TPX2 proteins.
First, Kern and coauthor Adelajda Hadzipasic along with colleagues at Brandeis, compared versions of Aurora A and TPX2 from a diverse set of species - including bacteria, plants, and humans.
Then, they did bioinformatic analysis, traveling back in time to deduce the ancient amino acid sequences of each protein and its oldest common ancestors. They then used those sequences to recreate the ancient proteins.
"Evolution used to be speculative subject, but it has now become 'experimental'. That's what is really exciting about this technique."
Christopher Miller, an HHMI Investigator alum and recently retired biochemist who used to work next door to Kern at Brandeis.
Performing experiments in the lab allowed Kern's team to map how and when the 2 proteins' initially entwined.
Around 1.5 billion years ago, Aurora A self-activated without TPX2. But a half billion years later TPX2 showed up and increased Aurora A's productivity by pulling it into the cellular job site.
"That created an evolutionary increase in the fitness of TPX2," explains Miller. Teaming up with TPX2 boosted Aurora A's efficiency as well, so their partnership endured.
The two proteins now had time to develop a sophisticated relationship, Kern adds. With time, TPX2 gained an additional role asactivator. Today, the modern Aurora A/TPX2 pairing works 10 times faster than Aurora A alone.
And when Kern's team combined ancient Aurora A with modern TPX2, they found the two proteins bound together surprisingly well - despite almost a billion years of evolution between them.
That wasn't expected. Conventional wisdom held that proteins that work together also evolve together - so a change in one partner is matched by a complementary change in the other. Researchers had thought modern TPX2 would likely have amassed too many changes to connect with such an ancient protein.
However, Kern's team solved this mystery. Sections of Aurora A and TPX2 — that touch — stayed essentially the same over 1 billion years. Even as other parts of these proteins morphed and shifted over time.
Neel Shah, a chemist at Columbia University believes investigating the details of such interactions could provide a springboard for creating new types of drug interactions. Though there's a long road between identifying proteins' allosteric sites and actually designing drugs that can take advantage of them, he believes "these findings could be a starting point."
Evolution of a kinase allosteric site
Enzyme activity is often regulated by conformational changes coupled to binding of an effector at an allosteric site, a feature especially important for enzymes involved in signaling cascades. Hadzipasic et al. studied the origins of allosteric regulation of Aurora A, a kinase involved in progression of the eukaryotic cell cycle. Aurora A is allosterically regulated through the binding of an effector protein named TPX2, which also targets the kinase to spindle microtubules. By reconstructing ancestor kinase sequences, they found that TPX2 bound to an early Aurora A but had very weak activation that was gradually strengthened by evolution of an allosteric network within the kinase. An evolutionary advantage from localizing the active protein at the mitotic spindle may have driven the development of this regulatory mechanism.
Abstract
A myriad of cellular events are regulated by allostery; therefore, evolution of this process is of fundamental interest. Here, we use ancestral sequence reconstruction to resurrect ancestors of two colocalizing proteins, Aurora A kinase and its allosteric activator TPX2 (targeting protein for Xklp2), to experimentally characterize the evolutionary path of allosteric activation. Autophosphorylation of the activation loop is the most ancient activation mechanism; it is fully developed in the oldest kinase ancestor and has remained stable over 1 billion years of evolution. As the microtubule-associated protein TPX2 appeared, efficient kinase binding to TPX2 evolved, likely owing to increased fitness by virtue of colocalization. Subsequently, TPX2-mediated allosteric kinase regulation gradually evolved. Surprisingly, evolution of this regulation is encoded in the kinase and did not arise by a dominating mechanism of coevolution.
Authors
Adelajda Hadzipasic, Christopher Wilson, Vy Nguyen, Nadja Kern, Chansik Kim, Warintra Pitsawong, Janice Villal, Yuejiao Zheng1 and Dorothee Kern.
Return to top of page.
| |
|
Mar 13 2020 Fetal Timeline Maternal Timeline News
 "Allosteric regulation is arguably the biggest evolutionary step in the development of higher organisms.” CREDIT Dorothee Kern PHD.
|



