|
|
Developmental Biology - COVID-19
COVID-19 Virus Epidemic Has Natural Origins
Analysis of public genome sequence data from SARS-CoV-2 and related viruses found no evidence that the virus was made in a laboratory or otherwise engineered...
The COVID-19 coronavirus epidemic has a natural origin, according to Scripps' Research analysis of public gene sequence data from SARS-CoV-2 and related viruses. Scientists found no evidence the virus was made in a laboratory or otherwise engineered.
The novel SARS-CoV-2 coronavirus emerged in the city of Wuhan, China, last year and has since caused a large scale COVID-19 epidemic, spreading to more than 70 other countries. It is the product of natural evolution, according to findings published March 17th in the journal Nature Medicine.
That analysis of genome sequence data from SARS-CoV-2 and related viruses, found no evidence the virus was made in a laboratory or otherwise engineered.
"By comparing the available genome sequence data for known coronavirus strains, we can firmly determine that SARS-CoV-2 originated through natural processes."
Kristian Andersen PhD, Associate Professor, Immunology and Microbiology, Scripps Research; and Corresponding Author on the paper.
Coronaviruses are a large family of viruses that can cause illnesses ranging widely in severity. The first known severe illness caused by a coronavirus emerged with the 2003 Severe Acute Respiratory Syndrome (SARS) epidemic in China.
A second outbreak of severe illness began in 2012 in Saudi Arabia with the Middle East Respiratory Syndrome (MERS).
On December 31 of 2019, Chinese authorities alerted the World Health Organization of an outbreak of a novel strain of coronavirus causing severe illness, subsequently named SARS-CoV-2.
As of February 20, 2020, nearly 167,500 COVID-19 cases have been documented, although many more mild cases have likely gone undiagnosed. The virus has killed over 6,600 people.
Shortly after the epidemic began, Chinese scientists sequenced the genome of SARS-CoV-2 and made that data available to researchers worldwide. This gene sequence data showed how Chinese authorities rapidly detected the epidemic and how the number of COVID-19 cases are increasing because of human to human transmission after a single introduction into the human population.
Andersen and collaborators at several other research institutions used this sequencing data to explore the origins and evolution of SARS-CoV-2 by focusing in on several tell-tale features of this virus. Their analysis revealing a genetic template for spike proteins referencing the protrusions on the outside of the virus that grab and penetrate human and animal cells:
(1) the Receptor-Binding Domain (RBD), a type of grappling hook that grips onto host cells
(2) the Cleavage Site, a molecular can opener allowing the virus to crack open and enter host cells.
Evidence for Natural Evolution
Scientists found the RBD portion of the SARS-CoV-2 evolved to effectively target a molecular feature on the outside of human cells called ACE2. This is a receptor involved in regulating blood pressure. The SARS-CoV-2 spike protein is so effective at binding to human cells, scientists conclude this is the result of natural selection and not gene manipulation.
Evidence for natural evolution is supported by data found on SARS-CoV-2's backbone - its molecular structure. If someone was looking to engineer a new coronavirus as a pathogen, they would have made it from the backbone of a virus known to cause illness. But, scientists find the SARS-CoV-2 backbone is substantially different from known coronaviruses and mostly resembles viruses found in bats and pangolins.
"These two features: mutations in the RBD portion of the spike protein and its distinct backbone, rule out laboratory manipulation as a potential origin for SARS-CoV-2" explains Andersen.
According to Josie Golding, epidemics lead at UK-based Wellcome Trust, the findings by Andersen and his colleagues are "crucially important to an evidence-based view to the rumors that have been circulating about the origins of the virus (SARS-CoV-2) causing COVID-19." Concluding the virus is a product of natural evolution, and ending any speculation about deliberate genetic engineering.
Possible Origins of the Virus
Based on their genomic sequencing analysis, Andersen and his collaborators concluded that the most likely origins for SARS-CoV-2 followed one of two possible scenarios.
In one, the virus evolved to become a pathogen through natural selection in a non-human host - then jumped to humans. This is how previous coronavirus outbreaks emerged. Humans contracted the virus after direct exposure to civets (SARS) and camels (MERS). Then researchers proposed bats as the most likely host for SARS-CoV-2 as it is very similar to a bat coronavirus.

Shi Zhengli, Virologist, Wuhan Institute of Virology, looked for SARS in the Asian bat population.
Scientific American
However, there are no documented cases of direct bat-human transmission, suggesting another intermediate host was more likely involved.
Both distinctive features of SARS-CoV-2's spike protein (1) the RBD portion that binds to cells and (2) the cleavage site that opens the virus up — had to have evolved into their current state before entering humans. This would allow the virus to rapidly infect humans, as the virus would have evolved features making it pathogenic and able to spread between people.
In another scenario, a non-pathogenic version of the virus jumped from an animal host into humans and then evolved to its current dangerous state. For instance, a coronavirus from a pangolin could have been transmitted to a human, either directly or through an intermediary host such as civets or ferrets.

Pangolins live in Nepal, Bhutan, India, Thailand, Myanmar and China. Found in a wide range of habitats, they are not aggressive, are toothless, solitary, and roll into a ball to avoid danger.
The distinct cleavage site characteristic of SARS-CoV-2 could have evolved within a human host undetected, prior to the epidemic. Researchers found the SARS-CoV-2 cleavage site is similar to cleavage sites in strains of bird flu which transmit easily between people.
SARS-CoV-2 could have evolved into a virulent cleavage site in human cells and quickly kicked off the current epidemic.
Study co-author Andrew Rambaut cautioned that it is difficult if not impossible to know at this point which of the scenarios is most likely. But, if SARS-CoV-2 entered humans from an animal source, it raises the probability of future outbreaks.
Abstract
In spite of their precipitous encounter with the environment, newborn infants cannot readily mount T helper type 1 (TH1) cell antibacterial and antiviral responses. Instead, they show skewing toward TH2 responses, which, together with immunoregulatory functions, are thought to limit the potential for inflammatory damage, while simultaneously permitting intestinal colonization by commensals1,2,3. However, these collective capabilities account for relatively few T cells. Here we demonstrate that a major T cell effector function in human newborns is interleukin-8 (CXCL8) production, which has the potential to activate antimicrobial neutrophils and ?? T cells. CXCL8 production was provoked by antigen receptor engagement of T cells that are distinct from those few cells producing TH1, TH2 and TH17 cytokines, was co-stimulated by Toll-like receptor signaling, and was readily apparent in preterm babies, particularly those experiencing neonatal infections and severe pathology. By contrast, CXCL8-producing T cells were rare in adults, and no equivalent function was evident in neonatal mice. CXCL8 production counters the widely held view that T lymphocytes in very early life are intrinsically anti-inflammatory, with implications for immune monitoring, immune interventions (including vaccination) and immunopathologies. It also emphasizes qualitative distinctions between infants' and adults' immune systems.
Authors
Kristian G. Andersen, Andrew Rambaut, W. Ian Lipkin, Edward C. Holmes and Robert F. Garry .
Acknowledgements
The authors thank all those who have contributed sequences to the GISAID database (https://www.gisaid.org/) and analyses to Virological.org (http://virological.org/). We thank M. Farzan for discussions, and the Wellcome Trust for support. K.G.A. is a Pew Biomedical Scholar and is supported by NIH grant U19AI135995. A.R. is supported by the Wellcome Trust (Collaborators Award 206298/Z/17/Z—ARTIC network) and the European Research Council (grant agreement no. 725422—ReservoirDOCS). E.C.H. is supported by an ARC Australian Laureate Fellowship (FL170100022). R.F.G. is supported by NIH grants U19AI135995, U54 HG007480 and U19AI142790.
Funding for the research was provided by the US National Institutes of Health, the Pew Charitable Trusts, the Wellcome Trust, the European Research Council, and an ARC Australian Laureate Fellowship.
Affiliations
Department of Immunology and Microbiology, The Scripps Research Institute, La Jolla, CA, USA
Kristian G. Andersen
Scripps Research Translational Institute, La
Jolla, CA, USA
Kristian G. Andersen
Institute of Evolutionary Biology, University of Edinburgh, Edinburgh, UK
Andrew Rambaut
Center for Infection and Immunity, Mailman School of Public Health of Columbia University, New York, NY, USA
W. Ian Lipkin
Marie Bashir Institute for Infectious Diseases and Biosecurity, School of Life and Environmental Sciences and School of Medical Sciences, The University of Sydney, Sydney, Australia
Edward C. Holmes
Tulane University, School of Medicine, Department of Microbiology and Immunology, New Orleans, LA, USA
Robert F. Garry
Zalgen Labs, Germantown, MD, USA
Robert F. Garry
Corresponding author
Correspondence to Kristian G. Andersen.
Ethics declarations
Competing interests
R.F.G. is co-founder of Zalgen Labs, a biotechnology company that develops countermeasures to emerging viruses.
Contributions
D.G. co-designed the study, undertook all experiments with human materials, evaluated the results and co-wrote the manuscript; P.F. co-designed the study, was attending physician to the clinical trial to which the study is annexed, provided human samples, evaluated clinical data and edited the manuscript; A.V. undertook the immunohistology; M.-L.M. undertook the animal model experiments; N.J.S. evaluated immunohistology and provided samples; K.C. designed and supervised the clinical trial to which the study is annexed and edited the manuscript; R.C. co-formulated the study as an annex to a clinical trial and edited the manuscript; N.K. co-supervised the analysis of pathology and edited the manuscript; A.H. co-designed the study, evaluated the results and co-wrote the manuscript.
Return to top of page.
| |
|
Mar 19 2020 Fetal Timeline Maternal Timeline News
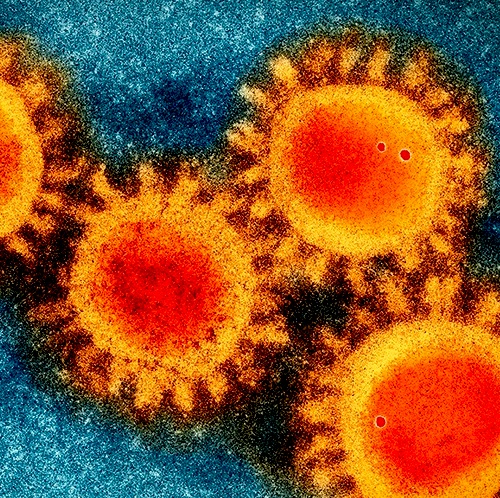 Electron microscopy photo of the COVID-19 coronavirus/
|





