|

CLICK ON weeks 0 - 40 and follow along every 2 weeks of fetal development
|
||||||||||||||||||||||||||||
|
Developmental Biology - Immune System Our Microbiome Signals Immune System Responses Epithelial tissue acts as an interface between our internal systems and our surrounding environment. Epithelial cells, therefore, are the gateway that pathogens must penetrate to gain access to our body and spread. In our body's defense, epithelial tissue has become colonized with a mix of bacteria, viruses, fungi and parasites collected over eons to combat these types of assaults. We call this matrix the microbiome. A team of researchers led by Andreas Diefenbach PhD, have been studying the microbiome's role coordinated with our body's immune response against harmful pathogens through the signaling pathways of each system. Infection triggers our body's immune response. A key role in this process is played by 'conventional dendritic cells' (cDCs). cDCs form part of a body's innate immune system and carry a range of pattern recognition receptors that enable quick detection of invading pathogens. • A cell's initial response involves release of cytokines, signaling proteins that attract immune cells to the site of infection. • At the same time, these cells use phagocytosis to engulf and digest invading pathogens. • After phagocytosis, cells produce individual particles on their surface as antigens. • This leads to activation of T cells (part of our adaptive immune system) and results in a targeted immune response to future assault by that pathogen. • In contrast, T cell activation triggered by cDCs from endogenous [originating from within our own body] antigens, leads to undesirable immune response and results in autoimmune diseases. Diefenbach's team found that cDCs cannot trigger immune attacks under sterile conditions (such as in germ-free mice). Thus, they conclude cDCs must receive information when in a 'basal (infection free) state, stimulated by microbiome signals. Interferons are cytokines, special signaling molecules known to play a role in antiviral activity. "Until now, we knew little about the role of IFN-I in a cell's basal state. cDCs, which do not receive IFN-I signaling during the basal state, cannot fulfill the physiological functions which they perform as part of the body's fight against pathogens," explains Diefenbach. Study results suggest the microbiome controls our immune system's fitness. It exerts this control by bringing the immune system to a state of 'readiness' in order to speed up its response to pathogens. Researchers used various animal models to study the manner in which the microbiome-controlled IFN-I primes basal-state cDCs for future combat. Using sequencing technology, they were able to compare the epigenomes and transcriptomes of cDCs from germ-free animals with those of control animals and animals deficient in IFN-I receptors. They wanted to know what happens at the molecular level in cDCs when they are no longer exposed to IFN-I. "Interestingly, when we looked at cDCs from germ-free animals and those without IFN-I signaling, we were able to observe low levels of expression among genes involved in the mitochondrial respiratory chain. This suggests that the microbiome is of crucial importance to the functioning of cDCs. It appears essential to the ability of cDCs to mount an effective response to bacterial or viral infections, including responses mediated by T cells. These findings may contribute to the development of new therapeutic approaches. Many autoimmune diseases, such as systemic lupus erythematosus, are caused by an increased production of IFN-I. Other studies have shown that the microbiome influences the effectiveness of checkpoint inhibitors in cancer immunotherapies. "These phenomena will continue to be of great interest to us. For instance, is it possible to change the composition of the microbiome in such a way as to reduce the availability of IFN-I, thereby exerting a positive influence on autoimmune diseases? Or might it be possible to improve responses to cancer immunotherapies by exerting a positive influence on the underlying IFN-I production?" explains Prof. Diefenbach. The team of researchers now plan to conduct further studies which will explore these questions. Summary Highlights • Microbiota controls constitutive IFN-I production by pDCs at steady state • Tonic IFNAR signaling instructs a basal state of cDCs, poised for future immune combat • Tonic IFN-I signals license cDCs for T cell priming against harmless peripheral antigens • Reduced metabolic fitness of Ifnar1 -/- cDCs cannot be overcome by direct activation Summary Environmental signals shape host physiology and fitness. Microbiota-derived cues are required to program conventional dendritic cells (cDCs) during the steady state so that they can promptly respond and initiate adaptive immune responses when encountering pathogens. However, the molecular underpinnings of microbiota-guided instructive programs are not well understood. Here, we report that the indigenous microbiota controls constitutive production of type I interferons (IFN-I) by plasmacytoid DCs. Using genome-wide analysis of transcriptional and epigenetic regulomes of cDCs from germ-free and IFN-I receptor (IFNAR)-deficient mice, we found that tonic IFNAR signaling instructs a specific epigenomic and metabolic basal state that poises cDCs for future pathogen combat. However, such beneficial biological function comes with a trade-off. Instructed cDCs can prime T cell responses against harmless peripheral antigens when removing roadblocks of peripheral tolerance. Our data provide fresh insights into the evolutionary trade-offs that come with successful adaptation of vertebrates to their microbial environment. Authors Laura Schaupp, Sabine Muth, Leif Rogell, Michael Kofoed-Branzk, Felix Melchior, Stefan Lienenklaus, Stephanie C. Ganal-Vonarburg, Matthias Klein, Fabian Guendel, Tobias Hain, Kristian Schütze, Ulrike Grundmann, Vanessa Schmitt, Martina Dorsch, Julia Spanier, Pia-Katharina Larsen, Thomas Schwanz, Sven Jäckel, Christoph Reinhardt, Tobias Bopp, Sven Danckwardt, Karsten Mahnke, Gitta Anne Heinz, Mir-Farzin Mashregh, Pawel Durek, Ulrich Kalinke, Oliver Kretz, Tobias B. Huber, Siegfried Weiss, Christoph Wilhelm, Andrew J. Macpherson, Hansjörg Schild, Andreas Diefenbach, Hans Christian Probst. Acknowledgements This study was funded by the European Research Council, University Ghent, Research Foundation Flanders (FWO), and the Health Research Council New Zealand. Research Partners and Funding Principal partners involved in the research were Prof. Dr. Hansjörg Schild, Dr. Hans Christian Probst and Dr. Sabine Muth of the Institute for Immunology/Research Center for Immunotherapy, University Medical Center Mainz. Other key partners were Prof. Dr. Stephanie Ganal-Vonarburg and Prof. Dr. Andrew Macpherson in Bern. Dr. Mir-Farzin Mashreghi of the German Rheumatism Research Center Berlin (DRFZ) was responsible for RNA sequencing. Other important partners included Prof. Stefan Lienenklaus and Prof. Dr. Ulrich Kalinke of the Hanover Medical School (MHH). Epigenome analyses were performed in collaboration with Dr. Thomas Manke of the Max Planck Institute of Immunobiology and Epigenetics in Freiburg. Metabolic analyses were performed in collaboration with Dr. Christoph Wilhelm of the Institute of Clinical Chemistry and Clinical Pharmacology, University Hospital Bonn. The study received substantial funding from the European Research Council (A. Diefenbach) and the German Research Foundation (A. Diefenbach, H.C. Probst and H. Schild). Return to top of page. | May 12 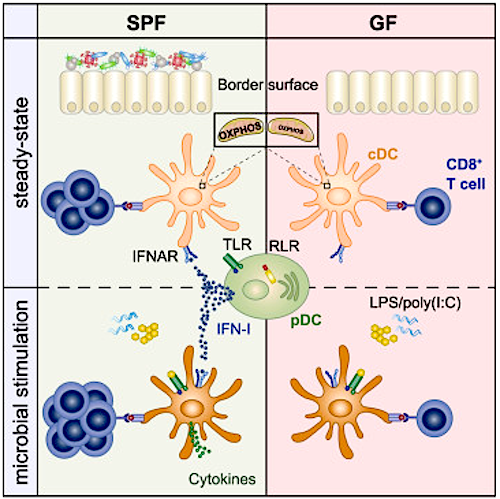 Environmental signals help shape our physiology and fitness. Centered in GREEN: Immune Cell CREDIT The Authors.
|
||||||||||||||||||||||||||||

