|
|
Developmental Biology - Sex Chromosomes
X and Y Chromosomes Need PAR
Breaking up is hard to do - especially for X and Y chromosomes...
The female X chromosome is large, containing thousands of genes critical to life. But the male Y chromosome is little, not more than a stub, yet must initiate male development and make sperm.
These two very different sized chromosomes must work together in order to pair-up during meiosis (a special form of cell division creating sperm and egg) and continue working together to make a baby.
How they pair-up has been mysterious for decades. But now scientists at the Sloan Kettering Institute have an idea of how they pair. It involves some very deliberate breaking and rejoining of DNA strands.
During meiosis, every chromosome from our mother lines up with every chromosome from our father — in order to swap segments.
But before swapping segments, chromosomal DNA must be broken so that "matching" or "homologous" regions on each chromosome can align. These 2 regions contain the same genes (but the particular DNA sequence on each may be slightly different).
Homologous recombination is vastly more challenging for males as most of the X chromosome has nothing to pair with on the tiny male chromosome.
In fact, only a very tiny portion of the already tiny Y chromosome has any homology with the X.
That tiny region is called the pseudoautosomal region (PAR), and is critical for X and Y chromosomes recombining into new sperm cells.
Scientists have known for a long time that the PAR region undergoes breaking and swapping of segments at a level far outpacing its size.
"On most chromosomes, DNA double-strand breaks typically occur once every 10 million base pairs. PAR in mice is less than 1/10 that size — but still undergoes frequent double-strand breaks."
Scott Keeney PhD, Molecular Biology Program, Memorial Sloan Kettering Cancer Center, New York; Louis V. Gerstner, Jr. Graduate School of Biomedical Sciences, Memorial Sloan Kettering Cancer Center, New York; and Howard Hughes Medical Institute, Maine, USA.
The research appears in a new study published May 27 in the journal Nature by Dr. Keeney and colleagues Laurent Acquaviva, research fellow, along with longtime collaborator Maria Jasin PhD a molecular biologist.
What's in a Blob?
The key to proper pairing of X and Y, they discovered, is a repeated sequence of DNA in the PAR that attracts several double-strand break-related proteins to this region. These protein clusters - which Dr. Acquaviva dubbed "blobs" - change the architecture of the chromosome in this region in such a way that the PAR becomes, as the authors put it, "the hottest area of double-strand break formation in the male mouse genome."
Similar blobs had been seen in images from published studies. But Laurent Acquaviva, a postdoctoral fellow in the Keeney lab and lead researcher on the project, and co-correspont author on the paper, was first to define what is is being seen in these blobs and connecting them to the hyper-accumulation of double-strand breaks in this region.
"At first glance, the blobs just look like a mess you might see in the microscope if the experiment didn't work. But they turned out to be completely predictable in number, timing, and location. So in reality they are very complex structures the cell builds on purpose."
Laurent Acquaviva PhD, Postdoctoral Researcher, Molecular Biology Program, Memorial Sloan Kettering Cancer Center, New York, NY, USA.
Acquaviva believes these blobs are PAR DNA tethered in short loops to the linear axis (structural backbone) of the chromosome.
Though the X chromosome has this same repeat DNA sequence, in meiosis neither female X chromosome typically recombines in this region. Why? SKI scientists show X chromosome pairings between regions happens first, and tends to directly opposes breakage at PAR.
This strategy of recruiting more than one's expected share of DNA-breaking proteins may not be limited to the PAR region. In a paper published earlier this month, the Keeney lab showed that small chromosomes in budding yeast resort to a similar tactic.
Groundbreaking Partnership
These new discoveries, which were made in mice, are the latest fruit of a longstanding collaboration between the Keeney and Jasin labs at SKI.
"Scott and I began collaborating back in 1997 when he joined SKI. This paper will be our 40th together. It's a tribute to the collaborative atmosphere of SKI," explains Maria Jasin PhD.
In fact, an accompanying editorial published along with the new paper was written by a former collaborative fellow, Francesca Cole, now a faculty member at MD Anderson Cancer Center. Drs. Jasin and Keeney are both interested in homologous recombination, and bring complementary expertise to their collaboration. Dr. Jasin is an expert in mammalian double-strand break repair, while Dr. Keeney is a specialist in yeast meiosis.
"I am hoping it [the latest research] will change the textbook version of how DNA strands move around during meiotic recombination. But many exciting questions still remain to be tackled."
Maria Jasin PhD; Louis V. Gerstner, Jr. Graduate School of Biomedical Sciences, Memorial Sloan Kettering Cancer Center, New York, NY; Developmental Biology Program, Memorial Sloan Kettering Cancer Center, New York, NY, USA.
Abstract
Sex chromosomes in males of most eutherian mammals share only a small homologous segment, the pseudoautosomal region (PAR), in which the formation of double-strand breaks (DSBs), pairing and crossing over must occur for correct meiotic segregation1,2. How cells ensure that recombination occurs in the PAR is unknown. Here we present a dynamic ultrastructure of the PAR and identify controlling cis- and trans-acting factors that make the PAR the hottest segment for DSB formation in the male mouse genome. Before break formation, multiple DSB-promoting factors hyperaccumulate in the PAR, its chromosome axes elongate and the sister chromatids separate. These processes are linked to heterochromatic mo-2 minisatellite arrays, and require MEI4 and ANKRD31 proteins but not the axis components REC8 or HORMAD1. We propose that the repetitive DNA sequence of the PAR confers unique chromatin and higher-order structures that are crucial for recombination. Chromosome synapsis triggers collapse of the elongated PAR structure and, notably, oocytes can be reprogrammed to exhibit spermatocyte-like levels of DSBs in the PAR simply by delaying or preventing synapsis. Thus, the sexually dimorphic behaviour of the PAR is in part a result of kinetic differences between the sexes in a race between the maturation of the PAR structure, formation of DSBs and completion of pairing and synapsis. Our findings establish a mechanistic paradigm for the recombination of sex chromosomes during meiosis.
Authors
Laurent Acquaviva, Michiel Boekhout, Mehmet E. Karasu, Kevin Brick, Florencia Pratto, Tao Li, Megan van Overbeek, Liisa Kauppi, R. Daniel Camerini-Otero, Maria Jasin and Scott Keeney .
Acknowledgements
The authors thank A. Tóth and B. de Massy for antibodies, mice, discussions and sharing of unpublished information; A. North and the Bio-Imaging Resource Center at Rockefeller University for assistance with SIM (supported by award number S10 RR031855 from the National Center For Research Resources); and R. Hendrickson, R. Soni and Z. Li (MSKCC Proteomics Core) for assistance with mass spectrometry. This work used the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov). MSKCC core facilities are supported by a Cancer Center Support Grant (P30 CA008748). L.A. was supported in part by a fellowship from the Lalor Foundation; M.B. was supported in part by a Rubicon fellowship from the Netherlands Organization for Scientific Research; and M.v.O. was supported in part by an NIH fellowship (F32 GM096692). This work was supported by NIGMS grants R35 GM118092 (S.K.) and R35 GM118175 (M.J.).
L.A. designed and conducted all of the cytogenetic experiments presented and analysed the data; M.E.K. generated Ankrd31 mutant mice and anti-ANKRD31 antibodies; M.B. and M.E.K. provided Ankrd31 mutant mice and unpublished data; M.E.K. performed the immunoprecipitation for mass spectrometry and T.L. validated the ANKRD31-interacting proteins; K.B. and F.P. performed SSDS and analysed the data under the supervision of R.D.C.-O., with input from L.A. and S.K.; M.v.O. generated REC8 and REC114 antibodies; L.K. performed initial characterization and provided unpublished data on the PAR ultrastructure and cohesin enrichment; M.J. and S.K. designed and supervised the research, analysed the data and secured funding; L.A. and S.K. wrote the manuscript with input from M.J. All authors edited the manuscript.
The authors declare no competing interests.
Return to top of page.
| |
|
Jun 2 2020 Fetal Timeline Maternal Timeline News
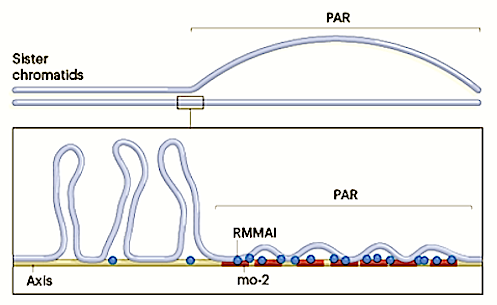 The PAR region of Male Y chromosome frequently undergoes double-strand breaks in order to swap DNA segments during meiosis. CREDIT Scott Keeney, Memorial Sloan Kettering.
|



