|
|
Developmental Biology - Hearing Development
Map of Mouse Inner Ear Sound Sensors
Valuable therapies for stem cell-based hearing loss...
A team of researchers has generated a developmental map of a key sound-sensing structure in the mouse inner ear.
Scientists at the National Institute on Deafness and Other Communication Disorders (NIDCD), part of the National Institutes of Health, with collaborators analyzed data from 30,000 cells from the mouse cochlea, a snail-shaped structure of the inner ear.
The results provide insights into genetic programs driving formation of cells important in detecting sounds. They also shed specific attention on the underlying cause of hearing loss linked to Ehlers-Danlos syndrome and Loeys-Dietz syndrome.
The study data is shared as a unique platform open to any researcher in an unprecedented resource catalyzing research on hearing loss.
Led by Matthew W. Kelley PhD, chief of Developmental Neuroscience at the NIDCD, the study is published online in Nature Communications. The research team includes investigators at the University of Maryland School of Medicine, Baltimore; Decibel Therapeutics, Boston; and King's College London.
"Unlike many other types of cells in the body, sensory cells that enable us to hear don't have the capacity regenerate when they become damaged or diseased.
Clarifying how these cells are formed in the developing inner ear, is an important asset for scientists working on stem cell-based therapeutics who need to treat or reverse forms of inner ear hearing loss."
Debara L. Tucci MD MS MBA, Otolaryngology-head and neck Surgeon; Director National Deafness and Other Communication Disorders, NIDCD.
In mammals, the primary transducers of sound are hair cells, spread across a thin ribbon of tissue (the organ of Corti) running the length of the coiled cochlea.
There are two kinds of hair cells, inner hair cells and outer hair cells, structurally and functionally sustained by several types of supporting cells. In development, a pool of nearly identical progenitor cells gives rise to these different cell types, but factors guiding transformation of progenitor cells into hair cells are not fully understood.
To learn how the cochlea forms, Kelley's team took advantage of a method called single-cell RNA sequencing. This powerful technique enables analysis of gene activity in a single cell. Scientists learn a lot about a cell from the gene patterns that encode proteins that define cell function. Patterns change during development in response to environmental cues.
"There are only a few thousand hair cells in the cochlea, arrayed close together in a complex mosaic arrangement making cells hard to isolate and characterize.
Single-cell RNA sequencing provides us with a valuable tool to track individual cell behavior as each cell takes its place in the intricate structure of the developing cochlea."
Michael E. Kelley PhD, Laboratory of Cochlear Development, National Institute on Deafness and Other Communication Disorders, National Institutes of Health, Bethesda, Maryland, USA.
Building on their earlier work on 301 cells, Kelley's team set out to examine gene activity profiles of 30,000 mouse cochleae cells. The cells were collected at specific time points: the 14th day of embryonic development — and the seventh postnatal day following birth. Collectively, the data is a vast catalog of information researchers used to explore cochlear development and study genes that underlie inherited forms of hearing impairment.
Kelley's team focused on one such gene, Tgf?r1, linked to two conditions associated in hearing loss:(1) Ehlers-Danlos syndrome and (2) Loeys-Dietz syndrome
Data showed Tgf?r1 is active in outer hair cell precursor cells as early as the 14th day of embryonic development. This suggests a gene that initiates formation of these cells.
To explore Tgf?r1's role, researchers blocked Tgf?r1 protein activity in cochleae from 14.5-day-old mouse embryos. When they examined these cochleae five days later, they saw fewer outer hair cells compared to embryonic mouse cochleae not treated with Tgf?r1 blocker. This finding suggests hearing loss in people with Tgf?r1 mutations could stem from impaired outer hair cell formation during development.
The study revealed other insights into early cochlear development. Pathways of inner and outer hair cells diverge early. Distinct gene activity appears at the earliest time, the 14th day of embryonic development, suggesting precursor cells which these cells derive from, are not as uniform as previously believed.
Additional research on cells collected at earlier stages is needed to characterize the initial steps in the formation of hair cells.
In the future, scientists may be able to use data to steer stem cells toward their hair cell lineage, helping to produce specialized cells in replacement approaches to revers some forms of hearing loss. The study's results also represent a valuable resource for research on the hearing mechanism and how it goes awry in congenital forms of hearing loss.
The authors made their data available through the gEAR portal (gene Expression Analysis Resource), a web-based platform for sharing, visualizing, and analyzing large multiomic datasets. The portal is maintained by Ronna Hertzano MD, PhD, and her team in the Department of Otorhinolaryngology and the Institute for Genome Sciences (IGS) at the University of Maryland School of Medicine.
"Single-cell RNA sequencing data are highly complex and typically require significant skill to access. By disseminating this study data via the gEAR, we are creating an 'encyclopedia' of the genes expressed in the developing inner ear, transforming the knowledge base of our field and making this robust information open and understandable to biologists and other researchers."
Hertzano
Significance
Anesthetics are used every day in thousands of hospitals to induce loss of consciousness, yet scientists and the doctors who administer these compounds lack a molecular understanding for their action. The chemical properties of anesthetics suggest that they could target the plasma membrane. Here the authors show anesthetics directly target a subset of plasma membrane lipids to activate an ion channel in a two-step mechanism. Applying the mechanism, the authors mutate a fruit fly to be less sensitive to anesthetics and convert a nonanesthetic-sensitive channel into a sensitive one. These findings suggest a membrane-mediated mechanism will be an important consideration for other proteins of which direct binding of anesthetic has yet to explain conserved sensitivity to chemically diverse anesthetics.
Abstract
Inhaled anesthetics are a chemically diverse collection of hydrophobic molecules that robustly activate TWIK-related K+ channels (TREK-1) and reversibly induce loss of consciousness. For 100 y, anesthetics were speculated to target cellular membranes, yet no plausible mechanism emerged to explain a membrane effect on ion channels. Here we show that inhaled anesthetics (chloroform and isoflurane) activate TREK-1 through disruption of phospholipase D2 (PLD2) localization to lipid rafts and subsequent production of signaling lipid phosphatidic acid (PA). Catalytically dead PLD2 robustly blocks anesthetic TREK-1 currents in whole-cell patch-clamp recordings. Localization of PLD2 renders the TRAAK channel sensitive, a channel that is otherwise anesthetic insensitive. General anesthetics, such as chloroform, isoflurane, diethyl ether, xenon, and propofol, disrupt lipid rafts and activate PLD2. In the whole brain of flies, anesthesia disrupts rafts and PLDnull flies resist anesthesia. Our results establish a membrane-mediated target of inhaled anesthesia and suggest PA helps set thresholds of anesthetic sensitivity in vivo.
Authors
Mahmud Arif Pavel, E. Nicholas Petersen, ProfileHao Wang, Richard A. Lerner and Scott B. Hansen.
Acknowledgements
This research was supported by the NIDCD Division of Intramural Research (ZIADC000039) as well as (ZICDC000086) to the Genomics and Computational Biology Core, which is led by Robert Morell, Ph.D., and by King's College London. The gEAR portal is primarily supported by the Hearing Restoration Project of Hearing Health Foundation, New York City, with additional funding from the NIDCD (R01DC013817) and from NIH's National Institute of Mental Health (R24MH114815). Computational resources of the NIH HPC Biowulf cluster were used in this study.
About the National Institute on Deafness and Other Communication Disorders (NIDCD): The NIDCD supports and conducts research and research training on the normal and disordered processes of hearing, balance, taste, smell, voice, speech, and language and provides health information, based upon scientific discovery, to the public.
About the National Institutes of Health (NIH): NIH, the nation's medical research agency, includes 27 institutes and centers and is a component of the U.S. Department of Health and Human Services. NIH is the primary federal agency conducting and supporting basic, clinical, and translational medical research, and is investigating the causes, treatments, and cures for both common and rare diseases.
The authors declare no competing interest.
Return to top of page.
| |
|
Jun 5 2020 Fetal Timeline Maternal Timeline News
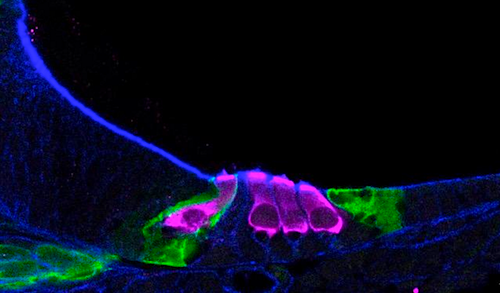 Single-cell RNA sequencing helped scientists map how sensory hair cells (pink) develop in a newborn mouse cochlea.
CREDIT Helen Maunsell, NIDCD/NIH.
|



