|
|
Developmental Biology - COVID 19
Detecting Antibodies to COVID19
Binding Domains may provide a specific target for detecting COVID19...
A new analysis of blood serum taken from 63 COVID-19 patients, 71 controls without COVID, and various coronavirus-exposed animals provides strong support for using the SARS-CoV-2 virus' receptor-binding domain (RBD) as an antigen — a macromolecule that causes an immune response — that would detect antibodies to COVID19.
The study appears in Science Immunology, and confirms the SARS-CoV-2 receptor-binding domain, or RBD, is capable of creating antibodies highly specific to SARS-CoV-2.
In fact, researchers found more than 95% of patients developed antibodies to a recombinant SARS-CoV-2 RBD antigen within nine days after their onset of symptoms.
Together, these results suggest RBD-based antigensor or receptor-binding domain, could be used to develop serological tests for SARS-CoV-2 exposure both specific for and sensitive to the SARS-CoV-2 virus.
The presence of RBD-induced antibodies strongly correlated with high levels of virus neutralizing antibodies in patients sampled.
Used in the general population, RBD-based antibody tests could provide an estimate of how many people have recovered from SARS-CoV-2 infections, a necessary first step for implementing policies to contain the pandemic and re-open communities.
Lakshmanane Premkumar and colleagues probed the antibody specificity of the SARS-CoV-2 RBD, a critical portion of the virus' spike protein complex, in sera sampled from symptomatic human patients, control patients, and animals exposed to various zoonotic coronaviruses.
They found that RBD antigen was 98% sensitive - it detected SARS-CoV-2 antibodies in 98% of individuals who tested positive for the virus via PCR tests - and 100% specific in the COVID-19 patient cohort, meaning that all patients who had tested positive for RBD-targeting antibodies were also positive for SARS-CoV-2 in PCR tests.
In some patients, RBD antigen cross-reacted with antibodies for SARS-CoV-1 (a related coronavirus that causes SARS), but as SARS-CoV-1 prevalence is very low in humans, this cross-reactivity is unlikely to pose diagnostic challenges, the scientists say.
Researchers plan to examine whether asymptomatic patients who carry the virus exhibit similar antibody responses as those people who experienced symptomatic infections.
Abstract
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) that first emerged in late 2019 is responsible for a pandemic of severe respiratory illness. People infected with this highly contagious virus can present with clinically inapparent, mild, or severe disease. Currently, the virus infection in individuals and at the population level is being monitored by PCR testing of symptomatic patients for the presence of viral RNA. There is an urgent need for SARS-CoV-2 serologic tests to identify all infected individuals, irrespective of clinical symptoms, to conduct surveillance and implement strategies to contain spread. As the receptor binding domain (RBD) of the spike protein is poorly conserved between SARS-CoVs and other pathogenic human coronaviruses, the RBD represents a promising antigen for detecting CoV-specific antibodies in people. Here we use a large panel of human sera (63 SARS-CoV-2 patients and 71 control subjects) and hyperimmune sera from animals exposed to zoonotic CoVs to evaluate RBD's performance as an antigen for reliable detection of SARS-CoV-2-specific antibodies. By day 9 after the onset of symptoms, the recombinant SARS-CoV-2 RBD antigen was highly sensitive (98%) and specific (100%) for antibodies induced by SARS-CoVs. We observed a strong correlation between levels of RBD binding antibodies and SARS-CoV-2 neutralizing antibodies in patients. Our results, which reveal the early kinetics of SARS-CoV-2 antibody responses, support using the RBD antigen in serological diagnostic assays and RBD-specific antibody levels as a correlate of SARS-CoV-2 neutralizing antibodies in people.
Authors
Lakshmanane Premkumar, Bruno Segovia-Chumbez, Ramesh Jadi, David R. Martinez, Rajendra Raut, Alena Markmann, Caleb Cornaby, Luther Bartelt, Susan Weiss, Yara Park, Caitlin E. Edwards, Eric Weimer, Erin M. Scherer, Nadine Rouphael, Srilatha Edupuganti, Daniela Weiskopf, Longping V. Tse, Yixuan J. Hou, David Margolis, Alessandro Sette, Matthew H. Collins, John Schmitz, Ralph S. Baric and Aravinda M. de Silva.
Acknowledgements
The authors gratefully acknowledge BEI Resources (https://www.beiresources.org) for the prompt processing and shipping of the reagents. We are grateful for the expert procedural care provided by the UNC Hospital and Blood Donor Center and to the patients and blood donors providing samples for the study. Funding: This work was funded by the University of North Carolina School of Medicine (L.P. and A.D.), National Institutes of Health Contract 75N9301900065 (A.S. and D.W.), NIH NIAID T32 AI007151 (D.M.) and a Burroughs Wellcome Fund Postdoctoral Enrichment Program Award (D.M.). Author contributions: Conceptualization: L.P., A.M.d S. Investigation: L.P., B.S., R.J., D.R.M., R.R., C.E.E. Resources: A.M, C.C, L.B., S.W., Y.P., E.W., E.M.S., N.R., S.E., D.W., L.V.T., Y.J.H., D.M., A.S., M.H.C., J.S., R.S.B. Writing: L.P., A.M.d S. Supervision: L.P, R.S.B, A.M.d S. Competing interests: The authors declare that they have no competing interests. Data and Materials Availability: The recombinant RBD antigens from the spike proteins used in this study are available under a standard MTA with the University of North Carolina. Please contact Lakshmanane Premkumar (prem@med.unc.edu) or Aravinda M. de Silva (aravinda_desilva@med.unc.edu). All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials. This work is licensed under a Creative Commons Attribution 4.0 International (CC BY 4.0) license, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited. To view a copy of this license, visit https://creativecommons.org/licenses/by/4.0/. This license does not apply to figures/photos/artwork or other content included in the article that is credited to a third party; obtain authorization from the rights holder before using such material.
Copyright © 2020, The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
This is an open-access article distributed under the terms of the Creative Commons Attribution license, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
Return to top of page.
|
|
Jun 16 2020 Fetal Timeline Maternal Timeline News
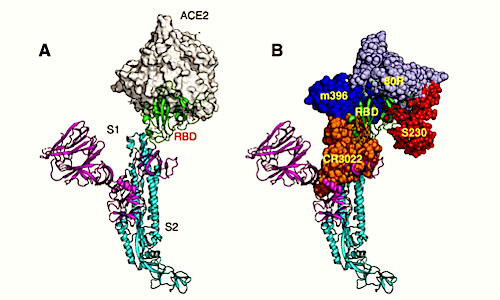 Production and characterization of RBD in coronavirus spike antigens. (A) The spike protein on the virion surface engages its Receptor Binding Domain or RBD. (B) RBD of the spike protein is the main human antibody target in SARS-CoV-1. (C) Amino acid sequence corresponding to RBD on the spike protein is poorly protected from SARS-CoV-2 and common human coronaviruses. (D) Coomassie-stained SDS-PAGE of purified spike RBD antigens from different CoVs. (E) Binding characterization of the spike RBD antigens with immune sera and a monoclonal antibody. SARS-CoV-1 monoclonal antibody (240C), serum from a mouse immunized with VRP expressing SARS-CoV-2 or SARS-CoV-1 spike protein, serum from a rabbit immunized with SARS-CoV-1 spike protein and an archived human sample collected before SARS-COV-2 were tested for binding against RBD spike antigens from SARS-CoV-2, SARS-Co-V-1, HCoV? (NL63) and HCoV? (HKU-1).
|
|

