|
|
Developmental Biology - Epigenetics
Individual Epigenetic Affects Identified
Epigenetic factors controlling all aspects of development can now be identified...
How does a fertilized egg cell develop into a complete organism with complex tissues and organs, although the gene information in each cell is exactly the same? A complex clockwork regulates how genes on a DNA molecule in each cell become active at the right time and place.
Scientists at the Max Planck Institute for Molecular Genetics in Berlin, Germany explored which factors affect embryonic development without altering DNA sequencing. Their results appear in the scientific journal Nature, describing how these mechanisms affect formation of tissues and organs in early mouse embryos.
Epigenetic regulation is part of the process of "packaging" DNA that modifies it without altering its underlying information. Specifically, epigenetic bookmarks attach to the DNA molecule and regulate which parts of the molecule are made active and in what order genes will become active.
Most epigenetic regulators, which are proteins, are essential to the embryo. Without them embryos die as organs begin developing. These regulators also have functions specific to particular cells — which makes them even more difficult to identify. How they affect embryo development is also relevant in cancer formations. The work appears in the scientific journal Nature.
Detailed Examination of Embryos
"The same regulator is present in all cells, but can have very different tasks, depending on cell type and time of development."
Stefanie Grosswendt, Department of Genome Regulation, Max Planck Institute for Molecular Genetics (MPIMG), Berlin, Germany, and one of the first authors of the study.
Grosswendt along with colleague Helene Kretzmer in the Alexander Meissner lab at MPIMG, and Zachary Smith, Harvard University, USA, captured data specific to ten of the most important epigenetic regulators and their affects on embryo development. Using the CRISPR-Cas9 system, they removed genes coding for these epigentic regulators in fertilized eggs and then observed how removal affected embryo growth.
Approximately six to nine days following removal of a regulator, researchers began seeing anatomical and molecular changes specific to each epigenetic regulator. Cells in many embryos were substantially altered. There were embryos with specific cells in excess, while other cells stopped dividing altogether.
Analyzing Thousands of Individual Cells
To make sense of these changes on a molecular level, researchers examined hundreds to thousands of individual cells from embryos, where one single epigenetic regulator was removed. They then sequenced the RNA molecules of almost 280,000 individual cells to investigate the consequences of loss of that regulator. RNA relays information encoded on DNA, allowing the scientists to identity cell behavior.
In their analysis, they identified a phase of development when epigenetic regulators are particularly important. After comparing data of altered and unaltered embryos, they were able to identify dysregulated genes and cell types abnormally over or under produced. From this data, they deduced previously unknown functions of many regulators.
Complex Effects During Development
An eight-day-old mouse embryo looks a bit like a seahorse but without any organs. "From the outer appearance of an early embryo, you can often only guess which structures and organs will form and which won't," explains Helene Kretzmer, bioinformatician and Zachary Smith, biologist, both first authors of the publication. "Our sequencing allows for a much more precise and high resolution view."
Single-cell analysis gave a highly detailed view over the first nine days of mouse development. Often, switching off a single regulator led to ripple effects throughout the network of interacting genes, with many differentially activated or inactivated genes over the course of development.
Removing the epigenetic regulator Polycomb (PRC2) had a striking impact. "Without PRC2, the embryo looks egg-shaped and very small after eight and a half days, which is very unusual," explained Kretzmer. "We see vast changes in how DNA is packaged [to fit within the nucleus] that happens much earlier, long before the embryo develops morphological abnormalities."
Researchers found PRC2 is responsible for limiting germline progenitor cells - the cells that become sperm and eggs. Without PRC2, the embryo develops an excessive number of PRC2 cells, loses its shape, and dies after a short time.
Starting Point For Further Analyses
"With this combination of new technologies, we have addressed issues up in the air for 25 years. We now better understand how epigenetic regulators arrange the many different types of cells in our body. Our method let us investigate other factors such as transcription, growth factors, or even a combination of both. We can now observe very early developmental stages at a level of detail previously unthinkable. The work is only the first step for even more detailed investigations."
Alexander Meissner PhD, Department of Genome Regulation, Max Planck Institute for Molecular Genetics, Berlin, Germany; Broad Institute of MIT and Harvard, Cambridge and Department of Stem Cell and Regenerative Biology, Harvard University, Cambridge, MA, USA; and leader of the study.
Abstract
During ontogeny, proliferating cells become restricted in their fate through the combined action of cell-type-specific transcription factors and ubiquitous epigenetic machinery, which recognizes universally available histone residues or nucleotides in a context-dependent manner1,2. The molecular functions of these regulators are generally well understood, but assigning direct developmental roles to them is hampered by complex mutant phenotypes that often emerge after gastrulation3,4. Single-cell RNA sequencing and analytical approaches have explored this highly conserved, dynamic period across numerous model organisms5,6,7,8, including mouse9,10,11,12,13,14,15,16,17,18. Here we advance these strategies using a combined zygotic perturbation and single-cell RNA-sequencing platform in which many mutant mouse embryos can be assayed simultaneously, recovering robust morphological and transcriptional information across a panel of ten essential regulators. Deeper analysis of central Polycomb repressive complex (PRC) 1 and 2 components indicates substantial cooperativity, but distinguishes a dominant role for PRC2 in restricting the germline. Moreover, PRC mutant phenotypes emerge after gross epigenetic and transcriptional changes within the initial conceptus prior to gastrulation. Our experimental framework may eventually lead to a fully quantitative view of how cellular diversity emerges using an identical genetic template and from a single totipotent cell.
Authors
Stefanie Grosswendt, Helene Kretzmer, Zachary D. Smith, Abhishek Sampath Kumar, Sara Hetzel, Lars Wittler, Sven Klages, Bernd Timmermann, Shankar Mukherji and Alexander Meissner.
Acknowledgements
The authors thank A. Bolondi, R. Weigert and other members of the Meissner laboratory, M. Chan and D. Hnisz for discussions and advice, S. Otto for experimental support characterizing the EED-knockout mES cell line, M. Walter for support with embryo isolations, T. Ahsendorf for help with initial efforts to optimize our genotyping pipeline, and D. Andergassen for discussions on SNP typing. We are also grateful to F. Koch and the transgenic facility, including M. Peetz, and C. Giesecke-Thiel of the Flow Cytometry Facility for their feedback and support. We thank M. Saitou for the mVenus Prdm14 promoter sperm that were provided by the RIKEN BRC through the National Bio-Resource Project of the MEXT/AMED, Japan (Acc. No. CDB0461T). This work was funded by the National Institutes of Health (NIH; 1P50HG006193, P01GM099117, 1R01HD078679 and 1DP3K111898) and the Max Planck Society.
Return to top of page.
|
|
Aug 7 2020 Fetal Timeline Maternal Timeline News
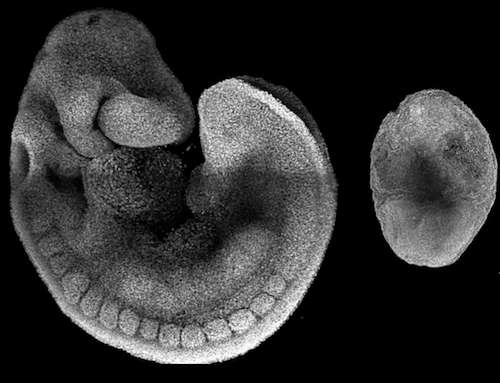 After 8 days, the developing mouse embryo resembles a seahorse [LEFT]. Without the epigenetic regulator PRC2, it is less complex and looks more like an egg [RIGHT]. A closer look into cells of each embryo reveals the tasks epigenetic regulatory factors affect during development. CREDIT Abhishek Sampath Kumar/ MPI
|
|



