|

CLICK ON weeks 0 - 40 and follow along every 2 weeks of fetal development
|
||||||||||||||||||||||||||||
|
Developmental Biology - Cell Signaling Two Discoveries Boost Next-Generation Organoids Implications for Tissue Engineering The sheer complexity of the new signaling roadmap helps explain why it took so long to make the initial three-organoid breakthrough. For example, the map revealed five distinct populations of mesenchymal cells involved in liver formation alone. Now, co-authors say the new roadmap will make the process faster, could expand the types of organs that can be grown together, and will allow researchers to grow sets of organoids engineered to mimic conditions that lead to birth defects or increased disease risk ± including some forms of cancer. "One important outcome of our study is the use of a signaling roadmap to direct the development of stem cells into different organ cell types. This approach may have important applications for tissue engineering." Short term, such organoid systems can be used to test new medications with far less dependence on animal models, or to evaluate the harms caused by pollution, unhealthy diets, allergens, and so on. Longer term, once expects learn ways to grow organoids to significantly larger sizes, customized lab-grown tissues could be used to repair damaged organs and someday even replace failing ones. Researchers layout in their paper protocols other scientists can use in making their own organoid systems. Detailed data also existes on their interactive website: Single cell transcriptomics of mouse foregut organogenesis. Abstract Visceral organs, such as the lungs, stomach and liver, are derived from the fetal foregut through a series of inductive interactions between the definitive endoderm (DE) and the surrounding splanchnic mesoderm (SM). While DE patterning is fairly well studied, the paracrine signaling controlling SM regionalization and how this is coordinated with epithelial identity is obscure. Here, we use single cell transcriptomics to generate a high-resolution cell state map of the embryonic mouse foregut. This identifies a diversity of SM cell types that develop in close register with the organ-specific epithelium. We infer a spatiotemporal signaling network of endoderm-mesoderm interactions that orchestrate foregut organogenesis. We validate key predictions with mouse genetics, showing the importance of endoderm-derived signals in mesoderm patterning. Finally, leveraging these signaling interactions, we generate different SM subtypes from human pluripotent stem cells (hPSCs), which previously have been elusive. The single cell data can be explored at: https://research.cchmc.org/ZornLab-singlecell. Authors Lu Han, Praneet Chaturvedi, Keishi Kishimoto, Hiroyuki Koike, Talia Nasr, Kentaro Iwasawa, Kirsten Giesbrecht, Phillip C. Witcher, Alexandra Eicher, Lauren Haines, Yarim Lee, John M. Shannon, Mitsuru Morimoto, James M. Wells, Takanori Takebe and Aaron M. Zorn. Acknowledgements This work was supported by grant NICHD P01HD093363 to A.M.Z. and J.W.M. T.T. is a New York Stem Cell Foundation – Robertson Investigator. M.M. research is supported by a Japanese Grant-in-Aid for Scientific Research (B) (17H04185). K.K. is supported by a Memorial Foundation postdoctoral fellowship and a RIKEN-CuSTOM collaborative grant for Promotion of Joint International Research (A) (18KK0423). This project was supported in part by NIH P30 DK078392 (Integrative Morphology, Sequencing and Pluripotent Stem Cell and Organoid Cores) of the Digestive Disease Research Core Center in Cincinnati. We are grateful to Minzhe Guo, Yan Xu, and Nathan Salomonis for help with the pseudotime and trajectory analysis, to James Briggs and Caleb Weinreb from the Alon Klein lab for advice on SPRING and members of the Rahul Satija lab for help with Seurat. Drs. Rafi Kopan and Emily Miraldi provided critical discussions and advice. Laboratory for Lung Development, RIKEN Center for Biosystems Dynamics Research (BDR), Kobe, 650-0047, Japan, Keishi Kishimoto and Mitsuru Morimoto CuSTOM-RIKEN BDR Collaborative Laboratory, Cincinnati Children’s Hospital, Cincinnati, OH, USA Keishi Kishimoto, Mitsuru Morimoto and Aaron M. Zorn CuSTOM, Division of Gastroenterology, Cincinnati Children’s Hospital, Department of Pediatrics, University of Cincinnati, College of Medicine, Cincinnati, OH, 45229, USA Hiroyuki Koike, Kentaro Iwasawa, Kirsten Giesbrecht and Takanori Takebe Division of Pulmonary Biology, Cincinnati Children’s Hospital, Department of Pediatrics, University of Cincinnati, College of Medicine, Cincinnati, OH, 45229, USA John M. Shannon Return to top of page. | Sep 1 2020 Fetal Timeline Maternal Timeline News 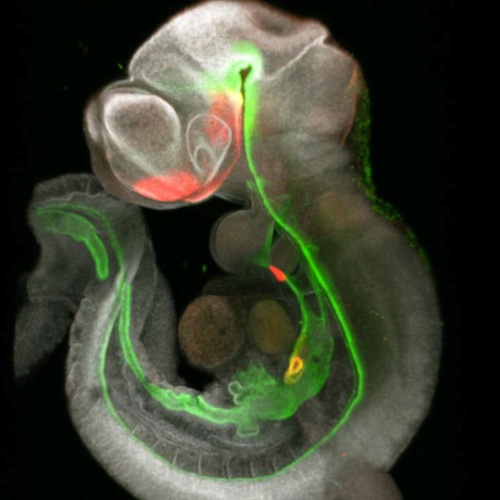 While this mouse foregut was forming, single-cell RNA sequencing captured 3 points of change from early cell patterning to induction of specific cell lines. CREDIT Cincinnati Children's and RIKEN
|
||||||||||||||||||||||||||||

