|
|
Developmental Biology - Memory and Sleep
Making Memories
Assembling neural activity into a finely tuned orchestra of memories — happens with the help of light and sleep...
Our memory is made up of impulses consolidated and stored in a matrix of proteins. Our brain stores these pulses in any number of engram cells — a unique cell system that translates neuronal activity into light, carefully discriminating between engram and non-engram cells.
Neural signals do this using proteins that fluoresce. Sub-ensembles of engram cells, represent distinct pieces of information constituting an entire memory. Some of these sub-ensembles appear during post-learning sleep, to be replayed. Some are more likely to be reactivated and retrieved while awake.
A Japanese research group supervised by Noriaki Ohkawa, lecturer along with Professor Kaoru Inokuchi at the University of Toyama, has established a system for investigating the characteristics of neuronal ensembles as they acquire memories. The research appeared in Japan Science and Technology Agency (JST) and was also republished in Nature Communications.
We are exposed to many episodic events throughout our lives. Episodic memory itself, is processed in several brain regions, one being the hippocampus. One type of hippocampal episodic memory is made up of engram cells active during learning.
The activation or inhibition of engram cell ensembles can induce or inhibit memory retrieval.
These ensembles are the actual manifestation of one specific memory.
One episodic memory may be composed of several episodes — with each encoded by a specific substrate or engram sub-ensembly.
Defining the Problem
But, it has never been clear how engram cells reassemble to represent a specific event. Also, technical limitations made it difficult to distinguish the activity of engram cells from that of non-engram cells. How one episodic memory is consolidated into an engram cell ensemble, requires seeing how the entire activity of ALL engram and non-engram cells react at the same time.
Developing Tools
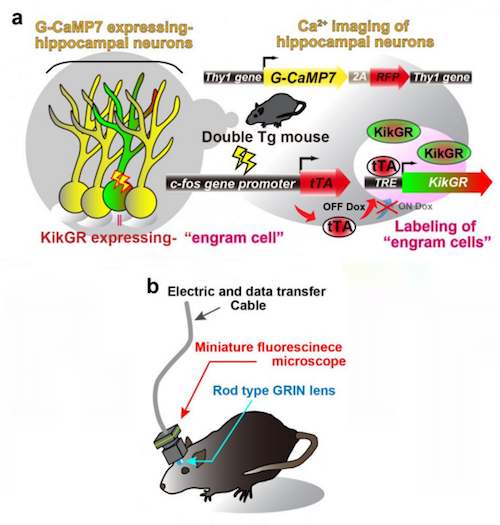
However, in c-fos-tTA rats engram cells can be specifically targeted. Their neural activity is in response to memory formation induced by the c-fos gene — which stimulates tTA expression.
• In the absence of doxycycline, tTA binds to tetracycline responsive element (TRE), enabling it to function as a TRE-dependent transgene (Fig. 1a).
• When a neuron is activated, Ca2+ flows into the soma and these rats express a Ca2+ indicator, G-CaMP7, in pyramidal neurons of their hippocampus.
• Neuronal activity is then seen as G-CaMP7 fluorescence or Ca2+ imaging.
Conducting the Experiment
Researchers soon developed a head-mounted, miniature fluorescent microscope to be mounted on the heads of double transgenic rats. Their goal being to look for Thy1-G-CaMP7/c-fos-tTA activity in these brains. Now, they could watch the hippocampal CA1 region and identify the Ca2+ signals corresponding to the activity of engram cells and non-engram cells, tracking novel episodic events (Figure 1c and 1d).
Forming A Theory
The data collected indicates that the activity of engram cell ensembles exhibit characteristic traits of highly repetitive activity during novel episodic events.
To address components of one memory, researchers deconstructed mousea population activity into sub-ensemble groups. Non-negative matrix factorization (NMF) decomposes population activity into a time series of coactive neuronal ensembles (Fig. 2a). Each sub-ensemble is composed of the different cells, which are spatially intermingled (Fig. 2b left), making their activity synchronous even among groups of engram cells associated with a single event (Fig. 2b right).
These results suggest the total information of one event is structured into sub-engram ensembles.
Testing that Theory
To measure the activity of engram cells across different memory processing stages, researchers retrieved and recorded Ca2+ moving from novel experience through post-experience sleep.
Around 40% of the total number of engram cell sub-ensembles appeared as novel experiences, reactivated during post-experience sleep and then preferentially reappeared in retrieval sessions. Almost none of those sub-ensembles in non-engram cells showed this feature.
Thus, engram sub-ensembles formed during a novel experience — and reactivated during sleep sessions — were mostly reactivated during the retrieval session (Fig. 3). In contrast, most non-engram ensembles activated during the novel experience were unable to be reactivated in later sessions.
Conclusion
Therefore, engram cells reflect synchronized activity via several sub-ensembles in the engram cell population.
Only engram cells perform this synchronous activity, surviving through post-learning sleep sessions — which contributes to their consolidation.
Abstract
The brain stores and recalls memories through a set of neurons, termed engram cells. However, it is unclear how these cells are organized to constitute a corresponding memory trace. We established a unique imaging system that combines Ca2+ imaging and engram identification to extract the characteristics of engram activity by visualizing and discriminating between engram and non-engram cells. Here, we show that engram cells detected in the hippocampus display higher repetitive activity than non-engram cells during novel context learning. The total activity pattern of the engram cells during learning is stable across post-learning memory processing. Within a single engram population, we detected several sub-ensembles composed of neurons collectively activated during learning. Some sub-ensembles preferentially reappear during post-learning sleep, and these replayed sub-ensembles are more likely to be reactivated during retrieval. These results indicate that sub-ensembles represent distinct pieces of information, which are then orchestrated to constitute an entire memory.
Authors
Khaled Ghandour, Noriaki Ohkawa, Chi Chung Alan Fung, Hirotaka Asai, Yoshito Saitoh, Takashi Takekawa, Reiko Okubo-Suzuki, Shingo Soya, Hirofumi Nishizono, Mina Matsuo, Makoto Osanai, Masaaki Sato, Masamichi Ohkura, Junichi Nakai, Yasunori Hayashi, Takeshi Sakurai, Takashi Kitamura, Tomoki Fukai and Kaoru Inokuchi.
Acknowledgements
The authors thank S. Tsujimura for genotyping the transgenic rats, M. Nomoto for introducing the CNMF-E and all members of the Inokuchi laboratory at the University of Toyama and the Fukai laboratory at RIKEN CBS for support and valuable discussion. This work was supported by the JST PRESTO program JPMJPR1684 (N.O.) and the JST CREST program JPMJCR13W1 (K.I.), by the Japan Society for the Promotion of Science KAKENHI (Grant Numbers JP26640008 and JP16H04653 (N.O.), JP16H06276 (M.Os.), JP15H05723 (J.N.), JP18H02595 (T.S.), and JP23220009 and JP18H05213 (K.I.)), by a Grant-in-Aid for Scientific Research on Innovative Areas “Memory dynamism” (JP25115002 (K.I.) and JP26115504 (J.N.)), by “Willdynamics” (JP16H06401 (T.S.)), by “Artificial Intelligence and Brain Science” (JP17H06036 (T.F.)) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), by the Rotary Yoneyama memorial foundation (K.G.), by the Lotte Research Promotion Grant (N.O.), by the Naito Foundation (N.O.), by the Ichiro Kanehara Foundation (N.O.), by the Tamura Science and Technology Foundation (N.O.), by the Mitsubishi Foundation (K.I.), by the Uehara Memorial Foundation (K.G. and K.I.) and by the Takeda Science Foundation (support to N.O. and K.I.).
Ethics Declarations
Competing interests
Y.H. receives research funding from Fujitsu Laboratories Ltd. and Dwango. The other authors declare no competing interests.
Funding
Japan Science and Technology Agency, Japan Society for the Promotion of Science, Ministry of Education Culture Sports Science and Technology (MEXT), Rotary Yoneyama memorial Foundation, Lotte Research Promotion Grant, Naito Foundation.
Return to top of page.
| |
|
Sep 4 2020 Fetal Timeline Maternal Timeline News
 In this study, scientists demonstrate that hippocampal capacity for memory storage of new memories is maintained by continuous neurogenesis of hippocampal circuits. Observed in rats, a decrease or increase in neurogenesis, delayed or sped up recovery of memory. CREDIT the authors.
|



