|
|
Developmental Biology - Genetic Disorders
Robotics Help Advance Developmental Biology
The study of developmental biology is getting robot help...
Scientists are using a custom made robot to survey how mutations occur in regions of the genome affecting development. These regulatory regions aren't genes, but stretches of DNA called enhancers which determine how genes turn on and off during development. Researchers described their findings - and the robot they created - in the October 14th issue of the journal Nature.
"The real star is the robot. It was extremely creative engineering."
David L. Stern PhD, Group Leader, Howard Hughes Medical Institute, Janelia Research Campus, Ashburn, Virginia, USA.
The project was led by a former postdoc in Stern's lab, Justin Crocker, now a group leader at the European Molecular Biology Laboratory (EMBL) in Heidelberg, Germany. In the past, to study the role of regulatory regions in development, scientists examined the effects of just a few mutations at a time. But Crocker and Stern wanted to study hundreds of mutations - to determine the role of every position in a regulatory region.
These experiments required a complex preparation protocol to be repeated in exactly the same way across thousands of fruit fly embryos. Even slight inconsistencies in the preparation of samples could change the results.
So the pair worked with the Janelia Experimental Technology team, jET, to design and build a robot that could do the finicky work of preparing samples.
"The timing of many of these steps is critical. For us, the goal was to be able to remove the human as much as we could from the equation, to make the experiments very standardized."
Justin Crocker PhD, European Molecular Biology Laboratory, Heidelberg, Germany.
jET engineers followed Crocker around the lab, watching him carry out each step of the complex protocol, and even tried their hand at the process themselves. Then they developed creative solutions for each step, ultimately designing a robot that could translate the manual movements into automated steps.
It took several years and two failed prototypes before the team landed on a winning design: a toaster-sized contraption that can consistently preserve and stain hundreds of fly embryos in different stages of development.
Crocker and his colleagues used the Hybridizer Robot to study the effects of mutations in one enhancer the Stern lab had studied for decades. Past research suggested there was a lot of flexibility in the exact genetic sequence of this particular enhancer - lots of room for mutations to change the sequence without having big effects on function. But with the help of the Hybridizer, the new comprehensive survey presented a different picture.
Crocker's team created many variants of this enhancer, each with a handful of random mutations. Then they created 800 different fly strains, each carrying a different enhancer variant. Stern's lab has even made one-off modifications to this same enhancer in the past. But without the technological assist from the Hybridizer, it hadn't been possible to analyze the outcomes of hundreds of different possible changes.
Crocker and Stern initially thought this experiment would highlight a few small regions critical to the enhancer's function. But surprisingly, they found that most of the mutations altered gene expression [function] in some way. And it didn't matter where those mutations were made: almost all areas of the enhancer seemed to encode valuable information.
"The regulatory regions of genes encode information much more densely than previously appreciated. Both of these facts may constrain how regulatory regions evolve."
David L. Stern PhD
Single mutations often had several different effects - for example, reducing gene expression in some parts of the fly while increasing expression elsewhere.
Scientists aren't yet sure why that's the case or how their findings square with the fact that enhancers often evolve very quickly. "We hope it will lead people to start thinking about these regions differently and design new kinds of experiments to explore this problem more deeply," Stern says.
Crocker is now collaborating with engineers at EMBL to improve the Hybridizer and design new robotic tools, which he hopes will make it possible for others in his field to automate similar experiments. "It's allowing us to do entirely new types of experiments and answer questions that people haven't been able to touch before," Crocker adds.
Abstract
Changes in gene regulation underlie much of phenotypic evolution1. However, our understanding of the potential for regulatory evolution is biased, because most evidence comes from either natural variation or limited experimental perturbations2. Using an automated robotics pipeline, we surveyed an unbiased mutation library for a developmental enhancer in Drosophila melanogaster. We found that almost all mutations altered gene expression and that parameters of gene expression—levels, location, and state—were convolved. The widespread pleiotropic effects of most mutations may constrain the evolvability of developmental enhancers. Consistent with these observations, comparisons of diverse Drosophila larvae revealed apparent biases in the phenotypes influenced by the enhancer. Developmental enhancers may encode a higher density of regulatory information than has been appreciated previously, imposing constraints on regulatory evolution.
Authors
Timothy Fuqua, Jeff Jordan, Maria Elize van Breugel, Aliaksandr Halavatyi, Christian Tischer, Peter Polidoro, Namiko Abe, Albert Tsai, Richard S. Mann, David L. Stern, and Justin Crocker.
Acknowledgements
A.T. is supported by the German Research Foundation (Deutsche Forschungsgemeinschaft, TS 428/1-1) and EMBL. R.S.M. is supported by funding from R35GM118336. We thank J. Zaugg and J. Wirbel for statistical advice; C. J. Standley, X. Li, R. M. Galupa, G. A. Canales, M. Perkins and M. R. P. Alves for comments on the manuscript; C. Rastogi and H. Bussemaker for the NRLB algorithms; K. Richter and J. Sager for technical support; I. Jones for mounting the Drosophila species larvae; T. Laverty for Drosophila assistance; A. Milberger for help with CAD design; and J. Payne for discussions.
Return to top of page.
| |
|
Oct 21 2020 Fetal Timeline Maternal Timeline News
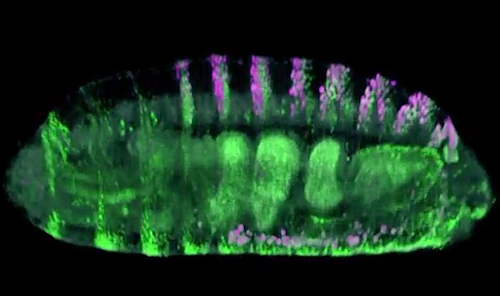 Researchers made hundreds of different mutations to an enhancer that acts on the "shavenbaby" fruit fly gene. They then tracked how those mutations altered when and where the gene was expressed [made to function]. In this fly embryo, shavenbaby is highlighted in GREEN, and the enhancer under study is highlighted in MAGENTA.
CREDIT Justin Crocker & Timothy Fuqua at EMBL
|



