|
|
Developmental Biology - Atlas of Molecular Function
Atlases Reveal How Cells Grow and Develop
Two atlases map gene function and how cells differentiate into cell types and unique tissues in humans...
Researchers at Brotman Baty Institute at the University of Washington (UW) Medicine in Seattle, have created two cell atlases that track gene expression and chromatin access during cell development. One atlas maps all gene expression within 15 individual fetal tissues. The second atlas maps chromatin accessibility for each individual gene within a cell.
Both atlases are fundamental to understanding gene and chromatin development at unprecedented scale. Techniques described making it possible to generate data for millions of cells.
Each atlas is described in two back-to-back articles, "A Human Cell Atlas of Fetal Gene Expression" and "A Human Cell Atlas of Chromatin Accessibility" in the November 13 issue of the journal Science
Gene Expression Atlas
Gene expression is when instructions stored in DNA are processed into proteins. Proteins determine a cell's structure and function, and each atlas captures where and when separate gene expression occurs and in each cell type as it develops.
"From these data, we can directly generate a catalog of all major cell types across human tissues, including how those cell types might vary in their gene expression in tissues."
Junyue Cao PhD, former postdoctoral fellow, laboratory of Jay Shendure; now Assistant Professor, Rockefeller University.
Once sequences were obtained, computer algorithms recovered single cell information, clustering cells by type and then subtypes, to identify developmental trajectories. The scientists profiled over 4 million single cells, identifying 77 main cell types and approximately 650 cell subtypes.
To create the Gene Expression Atlas, researchers profiled gene expression across 15 types of fetal tissue using a technique called sci-RNA-seq3. This technique labels each cell with a unique combination of three DNA "barcodes," allowing researchers to keep track of cells without ever physically separating them.
Researchers will next compare the new atlas to an existing atlas of mouse embryonic development. "When we combine this data with previously published data, we can directly delineate the cell's developmental path for all major cell types," explains co-senior author Cole Trapnell, associate professor, Genome Sciences, UW School of Medicine and a Brotman Baty Institute investigator.
DNA Accessibility Atlas
The second atlas of DNA accessibility, maps material called chromatin which allows DNA to be tightly wound and packed into a cell nucleus. Chromatin can be loosened and become "accessible" to molecular machinery reading encoded genetic instructions, or tighly wound and closed off, therefore "inaccessible" to molecular machinery.
Knowing regions of DNA that are open and closed can reveal how a cell chooses to turn genes on and off.
"Studying chromatin gives a sense of a cell's regulatory "grammar". Short stretches of DNA that are open, or accessible, are enriched with 'words' specifying certain genes be turned on."
Darren Cusanovich PhD, previously a postdoctoral fellow in the Shendure lab, now Assistant Professor, University of Arizona, and co-senior author.
In order to profile DNA accessibility within individual cells, researchers developed a method called sci-ATAC-seq3.
Similar to sci-RNA-seq3, the new technique also follows three DNA "barcodes" in each cell to tag and track individual cells. However, rather than identifying all the currently expressed sequences, sci-ATAC-seq3 captures and sequences only "open" chromatin sites.
In this study, researchers generated nearly 800,000 single-cell chromatin accessibility profiles at about 1 million sites across 15 fetal tissues.
They looked for proteins likely to interact with accessible DNA sites in each cell, and how those interactions explained specific cell types.
The research analysis defined control switches for development within the genome. Also identifying sites of chromatin accessibility that might be associated with diseases.
"This tells us what part of the genome might be functional. We still do not know what percentage of the genome that doesn't encode for genes can be involved in gene regulation. However, our atlas now provides that information for many cell types," explains Silvia Domcke, a co-lead author of the Accessibility Atlas paper and postdoctoral fellow in the Shendure lab.
Abstract
Introduction
A reference atlas of human cell types is a major goal for the field. Here, we set out to generate single-cell atlases of both gene expression (this study) and chromatin accessibility (Domcke et al., this issue) using diverse human tissues obtained during midgestation.
Rationale
Contemporary knowledge of the molecular basis of in vivo human development mostly derives from a combination of human genetics, in vivo investigations of model organisms, and in vitro studies of differentiating human cell lines, rather than through direct investigations of developing human tissues. Several challenges have historically limited the study of developing human tissues at the molecular level, including limited access, tissue degradation, and cell type heterogeneity. For this and the companion study (Domcke et al., this issue), we were able to overcome these challenges.
Results
We applied three-level single-cell combinatorial indexing for gene expression (sci-RNA-seq3) to 121 human fetal samples ranging from 72 to 129 days in estimated postconceptual age and representing 15 organs, altogether profiling 4 million single cells. We developed and applied a framework for quantifying cell type specificity, identifying 657 cell subtypes, which we preliminarily annotated based on cross-matching to mouse cell atlases. We identified and validated potentially circulating trophoblast-like and hepatoblast-like cells in unexpected tissues. Profiling gene expression in diverse tissues facilitated the cross-tissue analyses of broadly distributed cell types, including blood, endothelial, and epithelial cells. For blood cells, this yielded a multiorgan map of cell state trajectories from hematopoietic stem cells to all major sublineages. Multiple lines of evidence support the adrenal gland as a normal, albeit minor, site of erythropoiesis during fetal development. It was notably straightforward to integrate these human fetal data with a mouse embryonic cell atlas, despite differences in species and developmental stage. For some systems, this essentially permitted us to bridge gene expression dynamics from the embryonic to the fetal stages of mammalian development.
Conclusion
The single-cell data resource presented here is notable for its scale, its focus on human fetal development, the breadth of tissues analyzed, and the parallel generation of gene expression (this study) and chromatin accessibility data (Domcke et al., this issue). We furthermore consolidate the technical framework for individual laboratories to generate and analyze gene expression and chromatin accessibility data from millions of single cells. Looking forward, we envision that the somewhat narrow window of midgestational human development studied here will be complemented by additional atlases of earlier and later time points, as well as similarly comprehensive profiling and integration of data from model organisms. The continued development and application of methods for ascertaining gene expression and chromatin accessibility—in concert with spatial, epigenetic, proteomic, lineage history, and other information—will be necessary to obtain a comprehensive view of the temporal unfolding of human cell type diversity that begins at the single-cell zygote. An interactive website facilitates the exploration of these freely available data by tissue, cell type, or gene
Authors
Junyue Cao, Diana R. O’Day, Hannah A. Pliner, Paul D. Kingsley, Mei Deng, Riza M. Daza, Michael A. Zager, Kimberly A. Aldinger, Ronnie Blecher-Gonen, Fan Zhang, Malte Spielman, James Palis4, Dan Doherty, Frank J. Steemers, Ian A. Glass, Cole Trapnell and Jay Shendure.
Acknowledgements
Components of these studies were funded by the Brotman Baty Institute, the Paul G. Allen Frontiers Foundation, the Chan Zuckerberg Initiative, Hpward Hughes Medical Institute, and the National Institutes of Health.
Return to top of page.
| |
|
Nov 13 2020 Fetal Timeline Maternal Timeline News
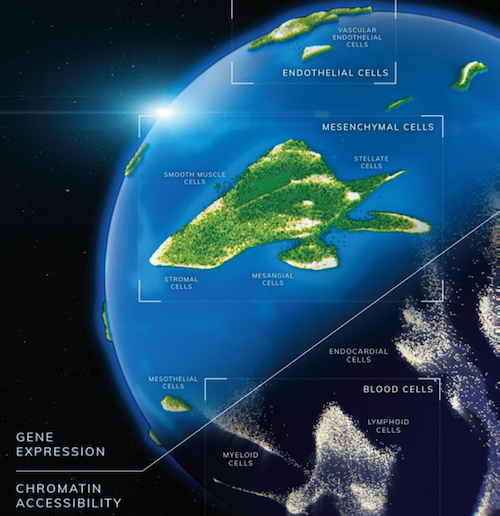 Artist's animation of two human cell atlases as they track gene expression and chromatin access during development of human cells and tissues. These cell atlases are based on research and technology by the Brotman Baty Institute and UW Medicine, Seattle. CREDIT Dani Bergey, Cognition Studios, Inc.
|



