|
|
Cell division errors: infertility, disorders and cancers
New insight into faulty cell formation helps us understand why congenital disorders such as Down's syndrome and perhaps infertility occur.
Researchers at the University of Edinburgh and the Max Planck Institute work on yeast — a model organism with cells similar to ours — in order to identify the key elements in preventing mistakes in cell division. Identifying cell division mistakes may aid our understanding of how infertility, stillbirths and birth defects arise as well. Being examined are the processes occurring just before cell division, essential to reproducing cells in the building of tissues.
The scientists observed a set of proteins known as Ctf19 which help suppress some aspects of the cell division re-organization process. Ctfl19 suppression helps accurately redistribute genes into packages we know as chromosomes. However, re-organization can misfire and lead to abnormal sections of DNA.
"Studying eggs and sperm in humans is very difficult for many reasons, but yeast provides a useful substitute from which we can learn a great deal. Our findings point to a process in which DNA is reorganised to ensure fertility."
Nadine Vincenten PhD, University of Edinburgh's School of Biological Sciences, who took part in the study.
The same error in DNA distribution in human eggs causes cells to have the wrong number of chromosomes — called aneuploidy — leading to congenital conditions such as Down syndrome.
The Ctf19 complex prevents DNA breaks and crossovers in both forms of cell division. However, in meiosis — when daughter cells divide to form haploid (half the chromosome number) of cells (as found in male or female sex cells or gametes) the division is initiated nearer the center of the centromere. Kinetochore spindles are known to assemble in the region of the centromere linking each chromosome to microtubule fibers during both mitosis and meiosis, that pull sister chromatids apart. But is was not known how critical the location within the centromere had to be.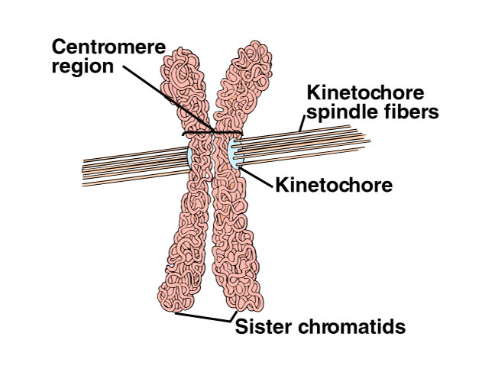
Using fluorescent markers, researchers highlighted how genetic material in yeast moved through cell division and saw when Ctf19 proteins are absent, cell division is more likely to be flawed.
In fact, researchers discovered multi-layered suppression by the Ctf19 complex operates only within the center of the centromere. Ctf19 complex prevents DNA crossovers during meiotic division and independently enhances cohesin — but only at the center of the centromere. It does not, however, prevent DNA breaks which initiate chromosome reorganization.
The discovery is a new non-standard role function for the kinetochore in defining chromosome domains. The kinetochore now seems to aid in preventing segregation errors that generate aneuploid gametes (male or female germ cells). These findings are published in the journal eLife.
Abstract
During meiosis, crossover recombination is essential to link homologous chromosomes and drive faithful chromosome segregation. Crossover recombination is non-random across the genome, and centromere-proximal crossovers are associated with an increased risk of aneuploidy, including Trisomy 21 in humans. Here, we identify the conserved Ctf19/CCAN kinetochore sub-complex as a major factor that minimizes potentially deleterious centromere-proximal crossovers in budding yeast. We uncover multi-layered suppression of pericentromeric recombination by the Ctf19 complex, operating across distinct chromosomal distances. The Ctf19 complex prevents meiotic DNA break formation, the initiating event of recombination, proximal to the centromere. The Ctf19 complex independently drives the enrichment of cohesin throughout the broader pericentromere to suppress crossovers, but not DNA breaks. This non-canonical role of the kinetochore in defining a chromosome domain that is refractory to crossovers adds a new layer of functionality by which the kinetochore prevents the incidence of chromosome segregation errors that generate aneuploid gametes.
This research was supported by the Wellcome Trust.
Return to top of page
|
|
|
Feb 19, 2016 Fetal Timeline Maternal Timeline News News Archive
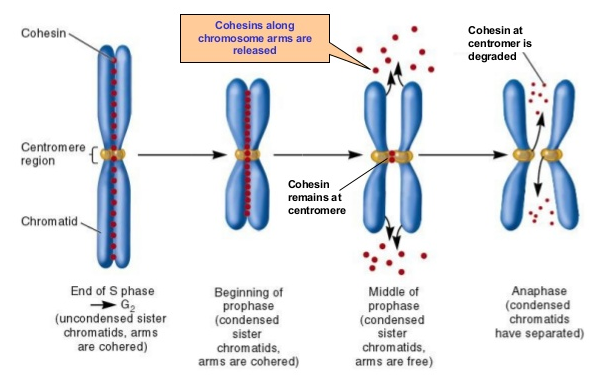
Chromosomes (blue) being pulled apart at centromere regions. Kinetochores (yellow) attach
spindle fibers and are then assisted by chohesin (red) in pulling sister chromatids apart.
Image Credit: McGraw Hill Publishers
|
|
|
|




